|
chr22_-_44258210
|
2.455
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chrX_-_13956530
|
2.351
|
|
GPM6B
|
glycoprotein M6B
|
|
chrX_-_13956445
|
2.319
|
|
|
|
|
chr14_+_96342728
|
2.306
|
|
LOC100507043
|
uncharacterized LOC100507043
|
|
chr12_+_41086189
|
2.209
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr8_+_104512975
|
2.193
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chrX_-_13956644
|
2.110
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr2_-_193059599
|
1.913
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr19_-_38714781
|
1.804
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr16_+_29912146
|
1.803
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr7_+_94285620
|
1.732
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr6_-_110500803
|
1.688
|
|
WASF1
|
WAS protein family, member 1
|
|
chr17_+_42836546
|
1.676
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr1_-_40157067
|
1.672
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr2_-_9143742
|
1.658
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr6_-_84418705
|
1.627
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr6_-_84418492
|
1.610
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr2_-_230579198
|
1.558
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr16_+_4838397
|
1.550
|
NM_001253790
NM_001253791
|
LOC440335
|
uncharacterized LOC440335
|
|
chr19_-_38720330
|
1.544
|
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr18_+_33877630
|
1.542
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr12_-_49392909
|
1.541
|
NM_015086
|
DDN
|
dendrin
|
|
chr6_+_56819923
|
1.509
|
|
BEND6
|
BEN domain containing 6
|
|
chr19_+_54385429
|
1.476
|
NM_002739
|
PRKCG
|
protein kinase C, gamma
|
|
chr12_+_79258448
|
1.436
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr3_-_187388105
|
1.434
|
NM_001048
|
SST
|
somatostatin
|
|
chr16_+_12995454
|
1.422
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr4_-_44450509
|
1.406
|
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chrX_-_51239426
|
1.389
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr8_-_22089850
|
1.378
|
NM_001099335
NM_014759
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr2_-_217236666
|
1.371
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr4_-_168155576
|
1.367
|
NM_001204353
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr17_-_39890882
|
1.363
|
NM_001079870
NM_001079871
NM_177977
|
HAP1
|
huntingtin-associated protein 1
|
|
chr19_-_51143091
|
1.356
|
NM_001160328
NM_001160329
|
SYT3
|
synaptotagmin III
|
|
chr16_+_2039945
|
1.351
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr11_-_46939908
|
1.351
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr10_+_64133915
|
1.304
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr8_+_35092963
|
1.299
|
NM_080872
|
UNC5D
|
unc-5 homolog D (C. elegans)
|
|
chr2_-_193059249
|
1.294
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr19_-_38720312
|
1.292
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr12_+_79258588
|
1.292
|
|
SYT1
|
synaptotagmin I
|
|
chr10_+_63422718
|
1.283
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr2_+_79740059
|
1.264
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr11_-_35440514
|
1.247
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr14_-_77607958
|
1.239
|
|
ZDHHC22
|
zinc finger, DHHC-type containing 22
|
|
chrX_+_53449804
|
1.238
|
NM_144968
NM_001031745
|
RIBC1
|
RIB43A domain with coiled-coils 1
|
|
chr17_-_37764165
|
1.228
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr17_+_64960754
|
1.224
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chrX_+_49126326
|
1.205
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr5_+_159656436
|
1.204
|
NM_001445
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr6_+_30851860
|
1.200
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr9_-_139948448
|
1.183
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr11_+_112832281
|
1.174
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr12_+_79258517
|
1.171
|
|
SYT1
|
synaptotagmin I
|
|
chr2_+_105471745
|
1.167
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr17_+_7554415
|
1.166
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr18_-_40695483
|
1.158
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chr19_-_47164289
|
1.156
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr9_-_122131579
|
1.145
|
NM_014618
|
DBC1
|
deleted in bladder cancer 1
|
|
chr10_-_101190201
|
1.138
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr17_+_48638339
|
1.135
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr20_+_44657858
|
1.119
|
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr5_-_114515733
|
1.111
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr8_-_75233461
|
1.111
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr1_-_109825767
|
1.103
|
NM_001005290
NM_001032291
NM_032636
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr8_+_24771276
|
1.101
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chrX_-_51239295
|
1.095
|
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr1_-_41950296
|
1.093
|
NM_001956
|
EDN2
|
endothelin 2
|
|
chr11_+_63871240
|
1.090
|
NM_013280
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1
|
|
chr1_+_70034029
|
1.088
|
|
LRRC7
|
leucine rich repeat containing 7
|
|
chr6_+_96463844
|
1.081
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr10_+_121485593
|
1.065
|
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr12_-_120806893
|
1.056
|
NM_002442
|
MSI1
|
musashi homolog 1 (Drosophila)
|
|
chr10_+_121485638
|
1.056
|
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr1_+_116916029
|
1.042
|
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr1_-_20812271
|
1.032
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr12_-_6809965
|
1.024
|
NM_001244014
NM_153685
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
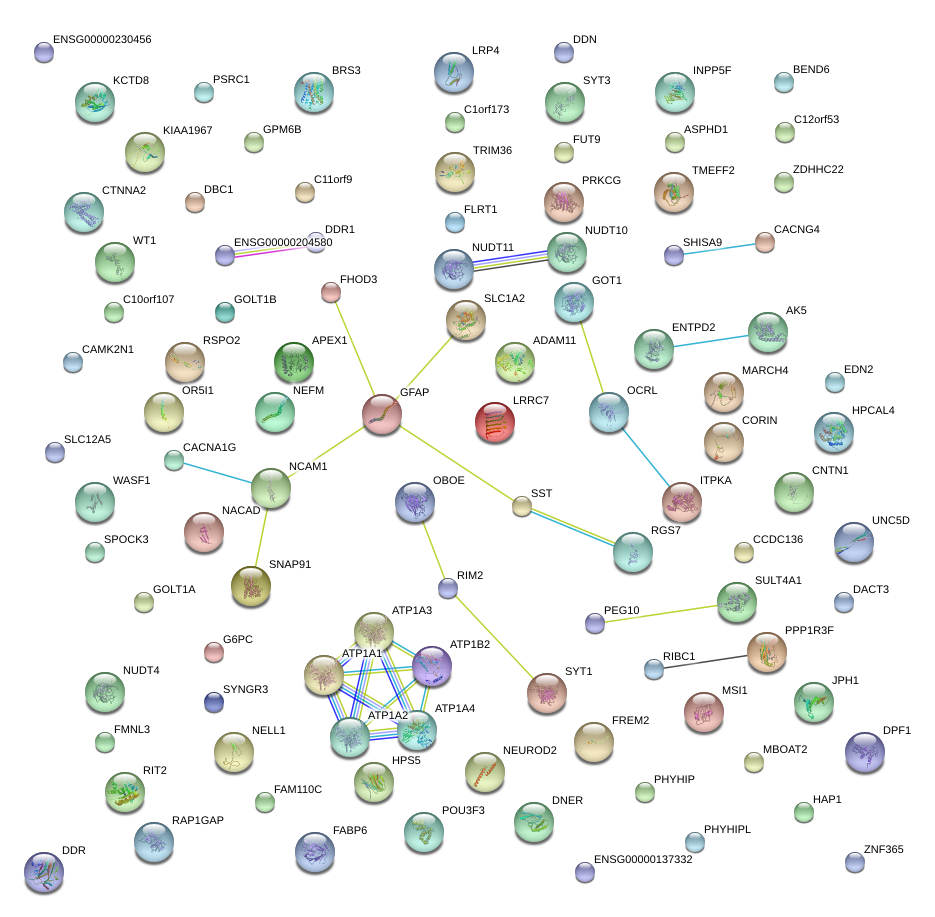
chr17_-_42988797
|
1.017
|
|
GFAP
|
glial fibrillary acidic protein
|
|
chr7_+_128432049
|
1.016
|
NM_022742
|
CCDC136
|
coiled-coil domain containing 136
|
|
chr15_+_41786055
|
1.007
|
NM_002220
|
ITPKA
|
inositol-trisphosphate 3-kinase A
|
|
chr1_-_75139421
|
1.000
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chrX_+_49126299
|
0.999
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chrX_+_135570124
|
0.995
|
NM_001727
|
BRS3
|
bombesin-like receptor 3
|
|
chr1_-_21995793
|
0.987
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr11_+_20691116
|
0.983
|
NM_006157
NM_201551
|
NELL1
|
NEL-like 1 (chicken)
|
|
chr1_-_109825683
|
0.979
|
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr7_-_45128492
|
0.978
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr1_+_77747727
|
0.968
|
|
AK5
|
adenylate kinase 5
|
|
chr2_+_79740171
|
0.964
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr8_+_24771268
|
0.957
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr8_-_109095836
|
0.956
|
NM_178565
|
RSPO2
|
R-spondin 2
|
|
chr15_-_23115243
|
0.950
|
|
LOC283683
|
uncharacterized LOC283683
|
|
chr13_+_39261172
|
0.946
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr2_-_46384
|
0.946
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr11_-_32457059
|
0.938
|
NM_000378
NM_024424
NM_024426
|
WT1
|
Wilms tumor 1
|
|
chr1_-_241520467
|
0.937
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr11_+_61522860
|
0.922
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr10_+_17686123
|
0.922
|
NM_003473
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1
|
|
chr22_-_44708730
|
0.917
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr19_+_12949258
|
0.916
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr8_-_22785228
|
0.912
|
NM_144962
|
PEBP4
|
phosphatidylethanolamine-binding protein 4
|
|
chr19_-_6502224
|
0.905
|
NM_006087
|
TUBB4A
|
tubulin, beta 4A class IVa
|
|
chr10_+_105036783
|
0.900
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr1_-_109825745
|
0.895
|
|
|
|
|
chr3_-_50540853
|
0.890
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr16_-_67427335
|
0.889
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr4_+_53728481
|
0.886
|
NM_023940
|
RASL11B
|
RAS-like, family 11, member B
|
|
chr3_-_133614421
|
0.885
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr18_-_24765178
|
0.884
|
NM_031422
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr13_-_102068705
|
0.881
|
NM_052867
|
NALCN
|
sodium leak channel, non-selective
|
|
chr11_+_12132063
|
0.878
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr1_-_32229635
|
0.878
|
NM_001703
|
BAI2
|
brain-specific angiogenesis inhibitor 2
|
|
chr8_-_22089517
|
0.878
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr1_+_109792640
|
0.877
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr22_-_46373007
|
0.876
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr8_-_30669934
|
0.866
|
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme
|
|
chr1_-_184006491
|
0.860
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr11_+_369723
|
0.857
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr1_+_161228516
|
0.856
|
NM_001102566
|
PCP4L1
|
Purkinje cell protein 4 like 1
|
|
chr3_-_47621729
|
0.856
|
NM_001206942
NM_001206945
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr14_-_23821423
|
0.850
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chrX_+_105969875
|
0.849
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr19_-_36523770
|
0.849
|
NM_015526
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr12_+_569542
|
0.847
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr6_-_32119717
|
0.846
|
NM_030651
|
PRRT1
|
proline-rich transmembrane protein 1
|
|
chr15_-_83240499
|
0.846
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr2_+_219824346
|
0.840
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr11_-_124622133
|
0.837
|
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr1_+_155829259
|
0.836
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr17_+_48610045
|
0.835
|
NM_017957
|
EPN3
|
epsin 3
|
|
chr19_-_7934806
|
0.834
|
|
FLJ22184
|
putative uncharacterized protein FLJ22184
|
|
chr20_+_24449834
|
0.832
|
NM_024893
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr11_+_17757494
|
0.830
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr12_-_22487566
|
0.827
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr9_+_124922189
|
0.827
|
NM_198469
|
MORN5
|
MORN repeat containing 5
|
|
chr13_+_35516390
|
0.825
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr12_-_130388211
|
0.825
|
NM_133448
|
TMEM132D
|
transmembrane protein 132D
|
|
chr5_+_173472692
|
0.819
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr16_+_58498182
|
0.819
|
|
NDRG4
|
NDRG family member 4
|
|
chr3_-_49907304
|
0.817
|
NM_024046
|
CAMKV
|
CaM kinase-like vesicle-associated
|
|
chr1_+_200863948
|
0.804
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr16_+_29823408
|
0.802
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr9_-_138591318
|
0.797
|
NM_001012415
NM_001101677
|
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1
|
|
chr1_-_223537400
|
0.796
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr1_+_77747602
|
0.789
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr8_+_69242956
|
0.788
|
NM_001195639
NM_052958
|
C8orf34
|
chromosome 8 open reading frame 34
|
|
chr4_-_21699317
|
0.784
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr12_-_6809595
|
0.784
|
NM_001244015
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
chr7_+_94536926
|
0.783
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr11_-_124622085
|
0.783
|
NM_014312
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chrX_+_18443702
|
0.782
|
NM_003159
|
CDKL5
|
cyclin-dependent kinase-like 5
|
|
chr8_+_85097099
|
0.782
|
NM_001100391
|
RALYL
|
RALY RNA binding protein-like
|
|
chr11_-_130298305
|
0.780
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr4_-_153700892
|
0.777
|
NM_145720
|
TIGD4
|
tigger transposable element derived 4
|
|
chr6_+_83903031
|
0.777
|
NM_033411
|
RWDD2A
|
RWD domain containing 2A
|
|
chr2_-_158184145
|
0.775
|
NM_001009959
|
ERMN
|
ermin, ERM-like protein
|
|
chr1_-_86622107
|
0.771
|
NM_152890
|
COL24A1
|
collagen, type XXIV, alpha 1
|
|
chrX_-_102319080
|
0.771
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr3_-_9290592
|
0.771
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr17_+_44068828
|
0.768
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr3_-_186079776
|
0.766
|
NM_001080744
NM_001080745
NM_001346
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr20_+_42875905
|
0.763
|
NM_024034
|
GDAP1L1
|
ganglioside-induced differentiation-associated protein 1-like 1
|
|
chr6_-_31846751
|
0.762
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chrX_-_139587224
|
0.761
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr14_-_25519076
|
0.761
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr2_-_51259536
|
0.761
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr6_+_72596405
|
0.756
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_-_73936299
|
0.754
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr11_+_119020572
|
0.750
|
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr12_-_62586547
|
0.749
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr16_+_84402125
|
0.746
|
NM_014861
|
ATP2C2
|
ATPase, Ca++ transporting, type 2C, member 2
|
|
chr9_-_34522972
|
0.741
|
NM_198573
|
ENHO
|
energy homeostasis associated
|
|
chr6_-_46138584
|
0.739
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr3_-_197024879
|
0.735
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr1_-_26232890
|
0.735
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chrX_+_14547419
|
0.734
|
NM_001118885
NM_001118886
NM_001171942
NM_002063
|
GLRA2
|
glycine receptor, alpha 2
|
|
chr2_-_139537803
|
0.732
|
NM_007226
|
NXPH2
|
neurexophilin 2
|
|
chr1_+_2005083
|
0.732
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr11_-_57414978
|
0.728
|
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr7_-_98467483
|
0.727
|
|
TMEM130
|
transmembrane protein 130
|
|
chr17_+_29718777
|
0.727
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chrX_-_128788913
|
0.726
|
NM_017413
|
APLN
|
apelin
|
|
chr15_+_43809805
|
0.724
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr1_+_16370246
|
0.723
|
NM_000085
|
CLCNKB
|
chloride channel Kb
|
|
chr14_+_79745653
|
0.723
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr17_+_20059192
|
0.722
|
NM_001033554
NM_001033555
NM_001243438
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr13_+_96743092
|
0.719
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr10_+_119806334
|
0.718
|
|
CASC2
|
cancer susceptibility candidate 2 (non-protein coding)
|
|
chr17_-_42988677
|
0.717
|
|
GFAP
|
glial fibrillary acidic protein
|
|
chr1_-_169396654
|
0.717
|
NM_021179
|
C1orf114
|
chromosome 1 open reading frame 114
|
|
chr5_-_78809658
|
0.716
|
NM_004272
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr5_+_173472737
|
0.713
|
|
HMP19
|
HMP19 protein
|
|
chr1_+_209757044
|
0.707
|
NM_020439
|
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG
|
|
chr2_-_175351755
|
0.705
|
NM_001033045
NM_152529
|
GPR155
|
G protein-coupled receptor 155
|
|
chr11_-_32456910
|
0.702
|
|
WT1
|
Wilms tumor 1
|
|
chr5_-_54529424
|
0.702
|
NM_021147
|
CCNO
|
cyclin O
|
|
chr6_+_72596648
|
0.698
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_-_63257545
|
0.697
|
NM_000524
|
HTR1A
|
5-hydroxytryptamine (serotonin) receptor 1A
|
|
chr1_+_155290639
|
0.695
|
NM_001105203
NM_001105204
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr1_-_202612580
|
0.692
|
NM_001136504
|
SYT2
|
synaptotagmin II
|