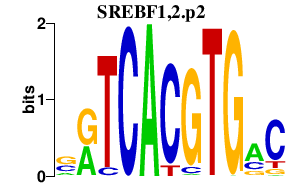
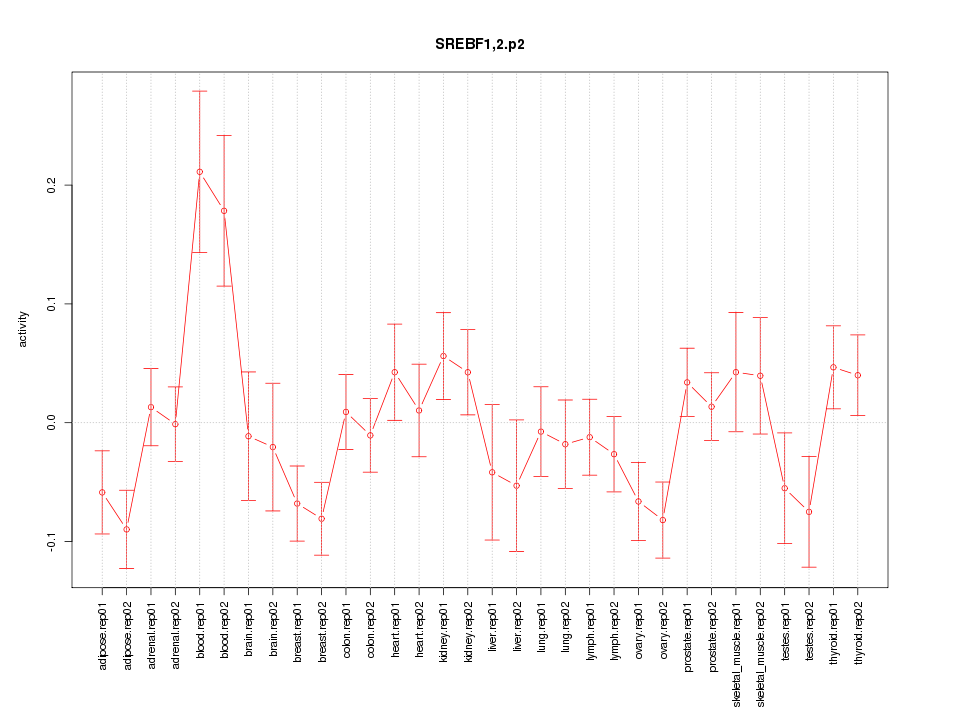
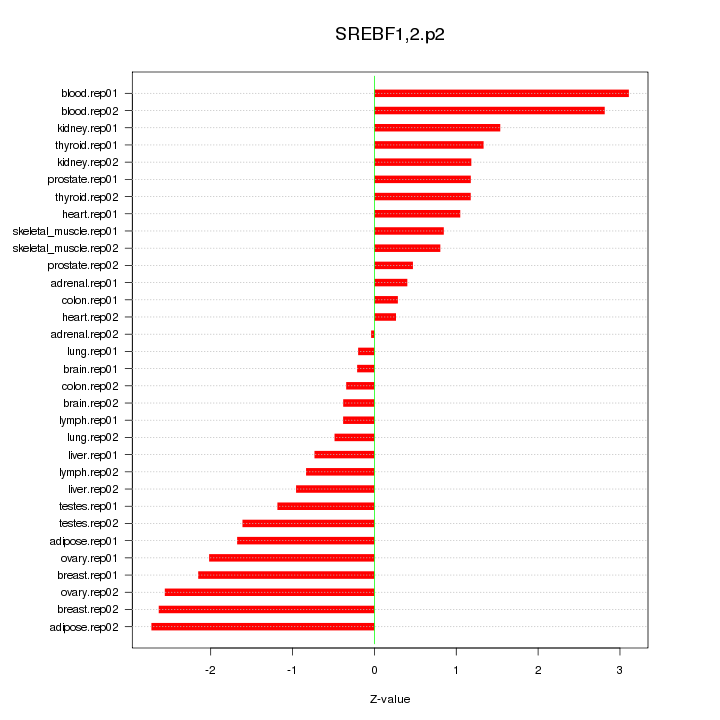
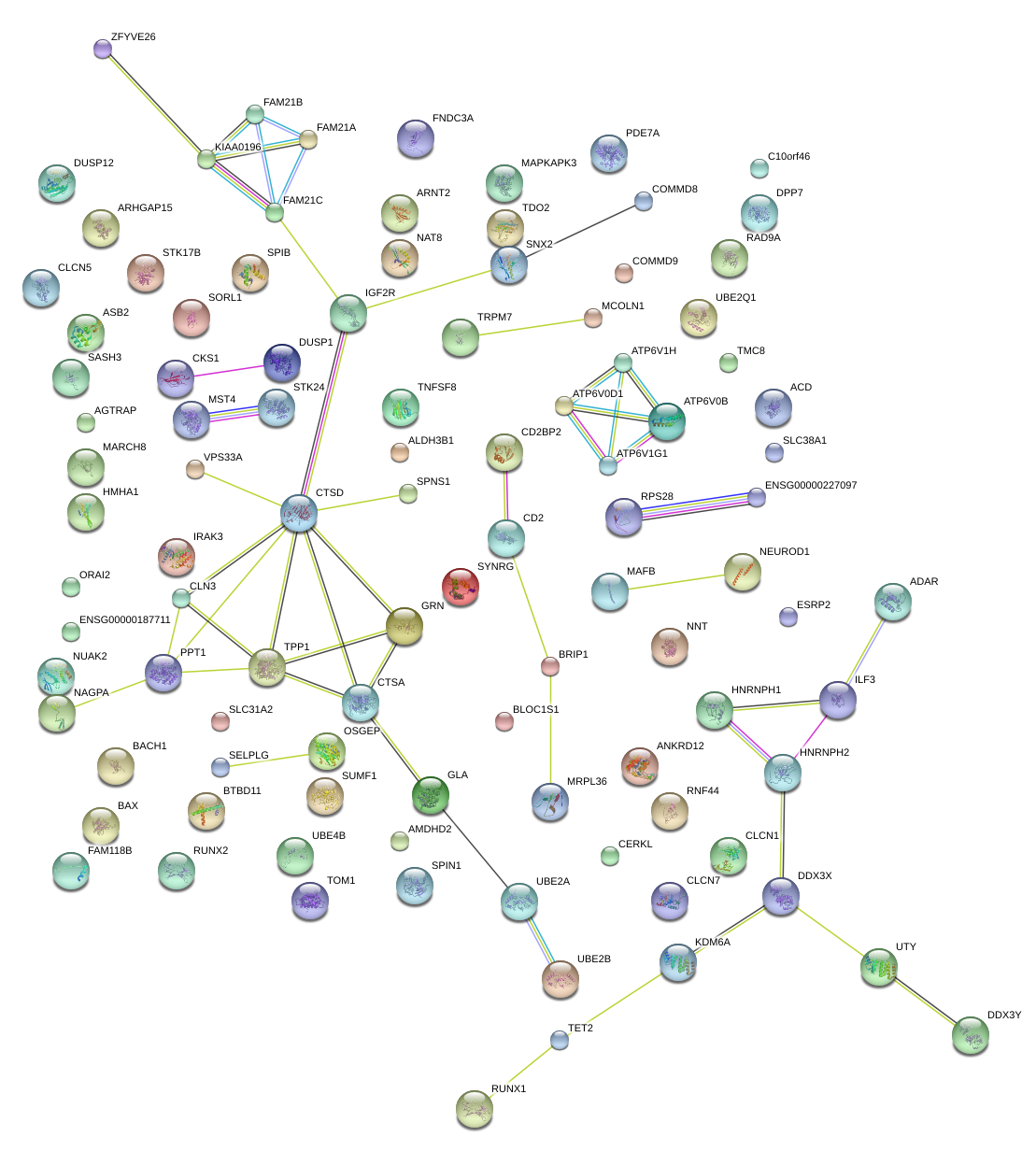
|
chr6_+_160390128
|
8.736
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr19_+_50922194
|
5.887
|
NM_001243998
NM_001243999
NM_001244000
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr19_+_49458031
|
4.387
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr11_+_67159421
|
4.299
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chrY_+_15016698
|
4.190
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr20_+_44519965
|
4.144
|
|
CTSA
|
cathepsin A
|
|
chr12_-_46662779
|
4.120
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr19_+_49458129
|
4.084
|
|
BAX
|
BCL2-associated X protein
|
|
chr1_+_44440673
|
3.961
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr20_+_44520203
|
3.944
|
|
CTSA
|
cathepsin A
|
|
chr11_-_36310934
|
3.882
|
|
COMMD9
|
COMM domain containing 9
|
|
chr7_+_102074002
|
3.858
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr19_+_49458151
|
3.801
|
|
BAX
|
BCL2-associated X protein
|
|
chr12_-_46662709
|
3.798
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr20_+_44519924
|
3.774
|
|
CTSA
|
cathepsin A
|
|
chr1_+_44440642
|
3.670
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr11_-_6640644
|
3.560
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr16_-_5083913
|
3.543
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr20_+_44520021
|
3.530
|
|
CTSA
|
cathepsin A
|
|
chr16_+_28986024
|
3.434
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr1_+_44440623
|
3.165
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr17_-_35969401
|
3.126
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr17_+_76126835
|
3.117
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr11_-_6640645
|
3.076
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr1_+_44440595
|
3.056
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr16_-_5083933
|
3.054
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr2_-_197036301
|
3.051
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr19_+_1067293
|
3.042
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr14_-_94443065
|
3.023
|
NM_001202429
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr19_+_10765106
|
2.979
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr5_+_43603312
|
2.950
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chrY_+_21729214
|
2.939
|
|
TXLNG2P
|
taxilin gamma 2, pseudogene
|
|
chr3_+_50654600
|
2.867
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr11_-_6640578
|
2.813
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr21_+_30671227
|
2.780
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr2_-_220042731
|
2.759
|
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1
|
|
chr20_+_44519590
|
2.721
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr11_-_71814268
|
2.721
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr1_-_154531083
|
2.705
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr12_+_56109851
|
2.703
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr3_+_50654558
|
2.670
|
NM_001243925
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr21_+_30671217
|
2.669
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr1_+_11796177
|
2.611
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr1_+_11796211
|
2.604
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr11_-_36310974
|
2.574
|
NM_001101653
NM_014186
|
COMMD9
|
COMM domain containing 9
|
|
chr3_+_50654589
|
2.553
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr2_-_197036262
|
2.534
|
|
STK17B
|
serine/threonine kinase 17b
|
|
chr20_-_39317875
|
2.524
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr8_-_54755836
|
2.522
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr5_+_133707105
|
2.501
|
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr1_+_11796128
|
2.494
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr19_+_7587436
|
2.448
|
NM_020533
|
MCOLN1
|
mucolipin 1
|
|
chr11_+_121323065
|
2.443
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr9_+_117349993
|
2.432
|
NM_004888
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chrX_+_131157635
|
2.421
|
NM_001042452
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr11_-_71814371
|
2.416
|
NM_017907
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr10_-_120514669
|
2.413
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr1_+_111742946
|
2.411
|
|
|
|
|
chr8_-_66701175
|
2.400
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chrX_-_100662788
|
2.397
|
|
GLA
|
galactosidase, alpha
|
|
chr1_-_40562923
|
2.382
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr4_-_47465719
|
2.382
|
|
COMMD8
|
COMM domain containing 8
|
|
chr12_-_109027654
|
2.380
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr3_+_50654868
|
2.375
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr13_+_49684388
|
2.370
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr17_+_42422490
|
2.350
|
NM_002087
|
GRN
|
granulin
|
|
chrY_-_15591799
|
2.330
|
|
UTY
|
ubiquitously transcribed tetratricopeptide repeat gene, Y-linked
|
|
chr11_+_121322848
|
2.306
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr8_-_54755546
|
2.302
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr12_-_109027615
|
2.294
|
|
SELPLG
|
selectin P ligand
|
|
chr16_-_67514989
|
2.290
|
NM_004691
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1
|
|
chr11_-_71814294
|
2.287
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr15_-_50978726
|
2.282
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr19_+_8386374
|
2.278
|
NM_001031
|
RPS28
|
ribosomal protein S28
|
|
chr14_-_68283293
|
2.272
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr12_-_122750954
|
2.270
|
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr9_+_115913227
|
2.270
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr16_-_28503622
|
2.263
|
NM_001042432
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chrY_+_15016018
|
2.249
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr16_+_2570322
|
2.249
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr4_+_106067031
|
2.249
|
NM_017628
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chrX_-_100662905
|
2.229
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr11_-_71814335
|
2.207
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr11_-_1785126
|
2.205
|
|
CTSD
|
cathepsin D
|
|
chr19_+_1067536
|
2.190
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr1_+_117297056
|
2.154
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr6_+_32936375
|
2.152
|
|
|
|
|
chr13_-_51417831
|
2.144
|
NM_198989
|
DLEU7
|
deleted in lymphocytic leukemia, 7
|
|
chr5_-_43603103
|
2.134
|
|
|
|
|
chr8_-_54755821
|
2.120
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr5_+_133706869
|
2.117
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr12_-_109027565
|
2.115
|
|
SELPLG
|
selectin P ligand
|
|
chr16_-_28503079
|
2.100
|
NM_000086
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr4_-_47465646
|
2.088
|
NM_017845
|
COMMD8
|
COMM domain containing 8
|
|
chr17_+_42422627
|
2.079
|
|
GRN
|
granulin
|
|
chr8_-_126103926
|
2.065
|
|
KIAA0196
|
KIAA0196
|
|
chrX_+_100663120
|
2.060
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_+_131157231
|
2.057
|
NM_001042453
NM_016542
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr5_+_43603216
|
2.050
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr9_+_117350097
|
2.046
|
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chr16_-_1525040
|
2.037
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr2_+_143886969
|
2.035
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr5_-_172198160
|
2.023
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr8_-_126104004
|
2.012
|
|
KIAA0196
|
KIAA0196
|
|
chr1_+_10092990
|
2.011
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B
|
|
chr14_-_20923266
|
2.008
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr5_-_175964223
|
1.993
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr18_+_9136742
|
1.990
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr3_-_4508952
|
1.985
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr22_+_35695872
|
1.971
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr16_-_68269948
|
1.965
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr7_+_143013218
|
1.962
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr21_-_36421558
|
1.955
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr10_-_120514455
|
1.949
|
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr1_-_205290756
|
1.945
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr16_-_1525006
|
1.942
|
|
CLCN7
|
chloride channel 7
|
|
chr9_-_140009171
|
1.899
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr16_-_1524926
|
1.887
|
|
CLCN7
|
chloride channel 7
|
|
chr11_+_126081618
|
1.884
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chrX_+_49832214
|
1.879
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr9_+_115913266
|
1.856
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr12_+_107974633
|
1.849
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr10_+_46222647
|
1.848
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
|
family with sequence similarity 21, member C
|
|
chrX_+_128913881
|
1.840
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr10_-_46090191
|
1.834
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr9_-_140009162
|
1.834
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr12_+_56109817
|
1.833
|
NM_001487
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr1_-_154600446
|
1.831
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr5_+_122110778
|
1.828
|
|
SNX2
|
sorting nexin 2
|
|
chr1_+_10093149
|
1.819
|
|
UBE4B
|
ubiquitination factor E4B
|
|
chr9_-_117692874
|
1.817
|
NM_001244
NM_001252290
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8
|
|
chr11_+_67776016
|
1.810
|
NM_001161473
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr12_+_66582914
|
1.806
|
NM_001142523
NM_007199
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chrX_+_128913897
|
1.800
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr8_-_126104007
|
1.797
|
|
KIAA0196
|
KIAA0196
|
|
chr2_-_182545212
|
1.795
|
NM_002500
|
CERKL
NEUROD1
|
ceramide kinase-like
neurogenic differentiation 1
|
|
chr17_-_76124767
|
1.787
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr16_-_1525020
|
1.785
|
|
CLCN7
|
chloride channel 7
|
|
chrX_+_100663213
|
1.779
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr9_-_140009169
|
1.772
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr12_-_122751044
|
1.767
|
NM_022916
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr1_-_40562909
|
1.765
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr1_-_153508597
|
1.759
|
NM_014624
|
S100A6
|
S100 calcium binding protein A6
|
|
chr21_+_38739193
|
1.757
|
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr17_+_78075360
|
1.755
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr8_-_126104021
|
1.753
|
|
KIAA0196
|
KIAA0196
|
|
chr1_+_110162427
|
1.753
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr3_-_187463227
|
1.751
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_-_15575333
|
1.749
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr11_+_60223281
|
1.745
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr3_-_187463474
|
1.742
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr10_+_23728197
|
1.738
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr5_-_131132690
|
1.736
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr21_+_30671113
|
1.729
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr10_+_69644708
|
1.723
|
|
SIRT1
|
sirtuin 1
|
|
chr10_-_3827205
|
1.712
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr20_+_43595176
|
1.702
|
|
STK4
|
serine/threonine kinase 4
|
|
chr9_-_140009194
|
1.693
|
NM_013379
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chrY_+_2709630
|
1.693
|
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr2_+_204571379
|
1.686
|
|
CD28
|
CD28 molecule
|
|
chr5_+_122110762
|
1.685
|
|
SNX2
|
sorting nexin 2
|
|
chr17_+_42422665
|
1.683
|
|
GRN
|
granulin
|
|
chr8_-_126103873
|
1.683
|
|
KIAA0196
|
KIAA0196
|
|
chr5_+_122110737
|
1.681
|
NM_003100
|
SNX2
|
sorting nexin 2
|
|
chr3_-_122512526
|
1.681
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr12_+_66583029
|
1.674
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr1_-_24513737
|
1.666
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr16_+_85061303
|
1.658
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr13_-_50159706
|
1.654
|
NM_018191
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr1_-_26393984
|
1.652
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr3_-_112280359
|
1.650
|
NM_022488
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chrX_+_100663229
|
1.649
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr1_+_15853348
|
1.647
|
NM_015291
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr10_+_69644341
|
1.643
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr11_-_1785104
|
1.643
|
|
CTSD
|
cathepsin D
|
|
chr2_+_220042986
|
1.635
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chrY_+_2803409
|
1.633
|
NM_001145275
NM_003411
|
ZFY
|
zinc finger protein, Y-linked
|
|
chr18_+_32556891
|
1.629
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr3_+_148709252
|
1.623
|
|
GYG1
|
glycogenin 1
|
|
chr19_+_1067114
|
1.619
|
NM_012292
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr10_+_51827654
|
1.619
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr8_-_126104045
|
1.615
|
NM_014846
|
KIAA0196
|
KIAA0196
|
|
chr7_-_29186072
|
1.602
|
NM_019029
NM_031311
|
CPVL
|
carboxypeptidase, vitellogenic-like
|
|
chr12_+_66583004
|
1.600
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr1_-_212873306
|
1.594
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr19_-_55895602
|
1.593
|
NM_001190764
|
TMEM238
|
transmembrane protein 238
|
|
chr22_+_35777087
|
1.591
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr17_-_47841517
|
1.587
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr1_-_186649421
|
1.576
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr7_+_102073953
|
1.572
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr11_-_85779821
|
1.571
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_+_24834083
|
1.569
|
NM_001251978
|
RCAN3
|
RCAN family member 3
|
|
chr14_+_20923341
|
1.566
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr12_-_46662533
|
1.561
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr8_+_133787556
|
1.557
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chrX_-_15873094
|
1.556
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr11_-_1785087
|
1.551
|
|
CTSD
|
cathepsin D
|
|
chr1_-_154928564
|
1.547
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr10_+_51827710
|
1.536
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr5_+_43603287
|
1.533
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|