|
chr19_-_45826131
|
74.254
|
|
CKM
|
creatine kinase, muscle
|
|
chr19_-_45826232
|
46.446
|
NM_001824
|
CKM
|
creatine kinase, muscle
|
|
chr2_+_74120132
|
43.077
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr15_-_35087748
|
42.367
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr1_-_229569839
|
40.271
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr2_+_74120092
|
39.327
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr20_+_35169876
|
32.651
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr14_-_94443065
|
29.473
|
NM_001202429
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr19_-_36247917
|
29.096
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr10_-_29923841
|
27.795
|
|
|
|
|
chr20_+_35169897
|
27.484
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr6_-_123957941
|
23.050
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr14_-_23877480
|
22.250
|
NM_002471
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha
|
|
chr17_+_33474835
|
21.367
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr14_-_94442894
|
21.043
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr8_+_86351055
|
20.480
|
NM_005181
|
CA3
|
carbonic anhydrase III, muscle specific
|
|
chr10_+_88428263
|
20.267
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr10_-_29923900
|
18.805
|
NM_021738
|
SVIL
|
supervillin
|
|
chr3_-_151987332
|
18.608
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr6_-_105584982
|
18.518
|
NM_001199563
|
BVES
|
blood vessel epicardial substance
|
|
chr1_-_16344438
|
17.631
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr2_+_88367298
|
17.619
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr1_-_208084379
|
16.589
|
|
CD34
|
CD34 molecule
|
|
chr1_-_208084432
|
16.379
|
NM_001025109
NM_001773
|
CD34
|
CD34 molecule
|
|
chr11_+_117070039
|
15.130
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr2_-_161056762
|
15.047
|
|
ITGB6
|
integrin, beta 6
|
|
chr22_+_31480981
|
14.925
|
NM_001207018
|
SMTN
|
smoothelin
|
|
chr20_-_42815732
|
14.831
|
|
JPH2
|
junctophilin 2
|
|
chr10_+_88428167
|
14.428
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr10_-_131762374
|
13.895
|
|
EBF3
|
early B-cell factor 3
|
|
chr3_-_46904879
|
13.010
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr1_-_242162364
|
12.910
|
NM_001004343
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma
|
|
chr12_+_52445185
|
12.802
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr11_+_66314373
|
12.675
|
NM_001104
|
ACTN3
|
actinin, alpha 3
|
|
chr3_-_99594915
|
12.562
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr1_+_16085245
|
12.368
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr4_-_88450601
|
11.999
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr18_-_3219846
|
11.029
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr10_-_90712510
|
10.565
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr5_+_42565963
|
10.470
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr4_+_24797084
|
10.469
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr2_-_128432642
|
10.216
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr7_-_27183225
|
9.958
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr2_-_106015423
|
9.831
|
NM_001039492
NM_001450
NM_201555
|
FHL2
|
four and a half LIM domains 2
|
|
chr15_-_65360297
|
9.478
|
NM_016563
|
RASL12
|
RAS-like, family 12
|
|
chr14_-_53417808
|
9.453
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr19_+_13135368
|
9.231
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_+_13134782
|
9.033
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr15_+_96869156
|
8.781
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_20306093
|
8.667
|
NM_001161729
|
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid)
|
|
chr5_-_33891967
|
8.395
|
NM_030955
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr17_+_41003184
|
7.740
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr4_-_88450243
|
7.407
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr7_-_107643330
|
7.369
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr17_-_1389323
|
7.178
|
|
MYO1C
|
myosin IC
|
|
chr6_+_151561094
|
7.103
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr15_+_96873845
|
7.032
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr5_+_42565562
|
6.688
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr3_+_187930720
|
6.430
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_-_154164551
|
6.347
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr7_+_20686965
|
6.298
|
NM_001163942
NM_001163993
NM_178559
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr16_-_11375094
|
5.759
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr18_-_3219986
|
5.746
|
NM_003803
NM_019856
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr12_+_26348488
|
5.670
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chrX_+_99839789
|
5.664
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chrX_-_153602928
|
5.624
|
|
FLNA
|
filamin A, alpha
|
|
chr17_+_32612686
|
5.418
|
NM_002986
|
CCL11
|
chemokine (C-C motif) ligand 11
|
|
chr20_+_4702499
|
5.411
|
NM_012409
|
PRND
|
prion protein 2 (dublet)
|
|
chr17_-_79792867
|
5.202
|
|
PPP1R27
|
protein phosphatase 1, regulatory subunit 27
|
|
chr5_-_33892292
|
5.171
|
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr7_+_114055048
|
5.052
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr11_-_65667809
|
5.009
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr10_+_75757847
|
4.944
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr6_+_151561508
|
4.920
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr17_-_10560625
|
4.707
|
NM_002470
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic
|
|
chr2_-_161056573
|
4.655
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr6_+_116832807
|
4.485
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr7_-_94285477
|
4.381
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr7_-_94285372
|
4.361
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr5_+_88185090
|
4.244
|
|
|
|
|
chr6_+_43138919
|
4.113
|
NM_003131
|
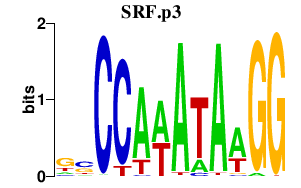
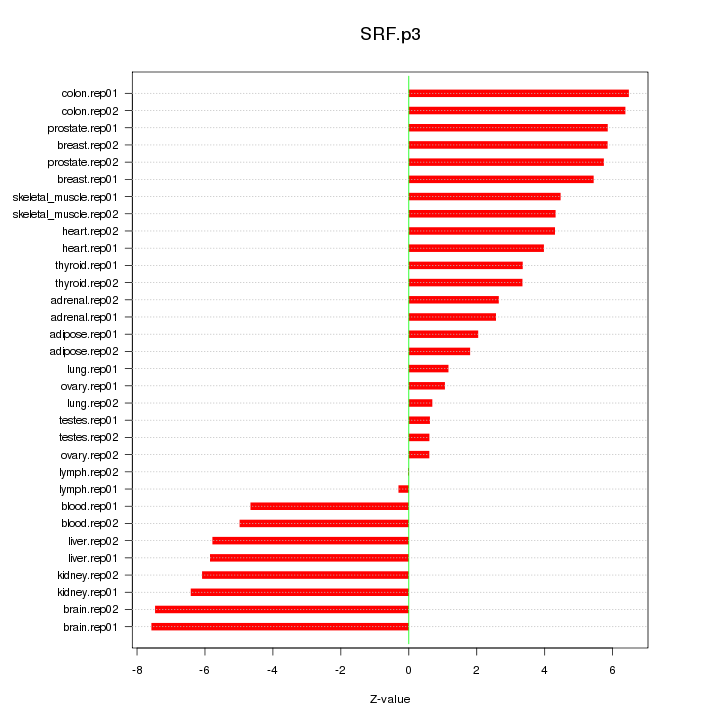
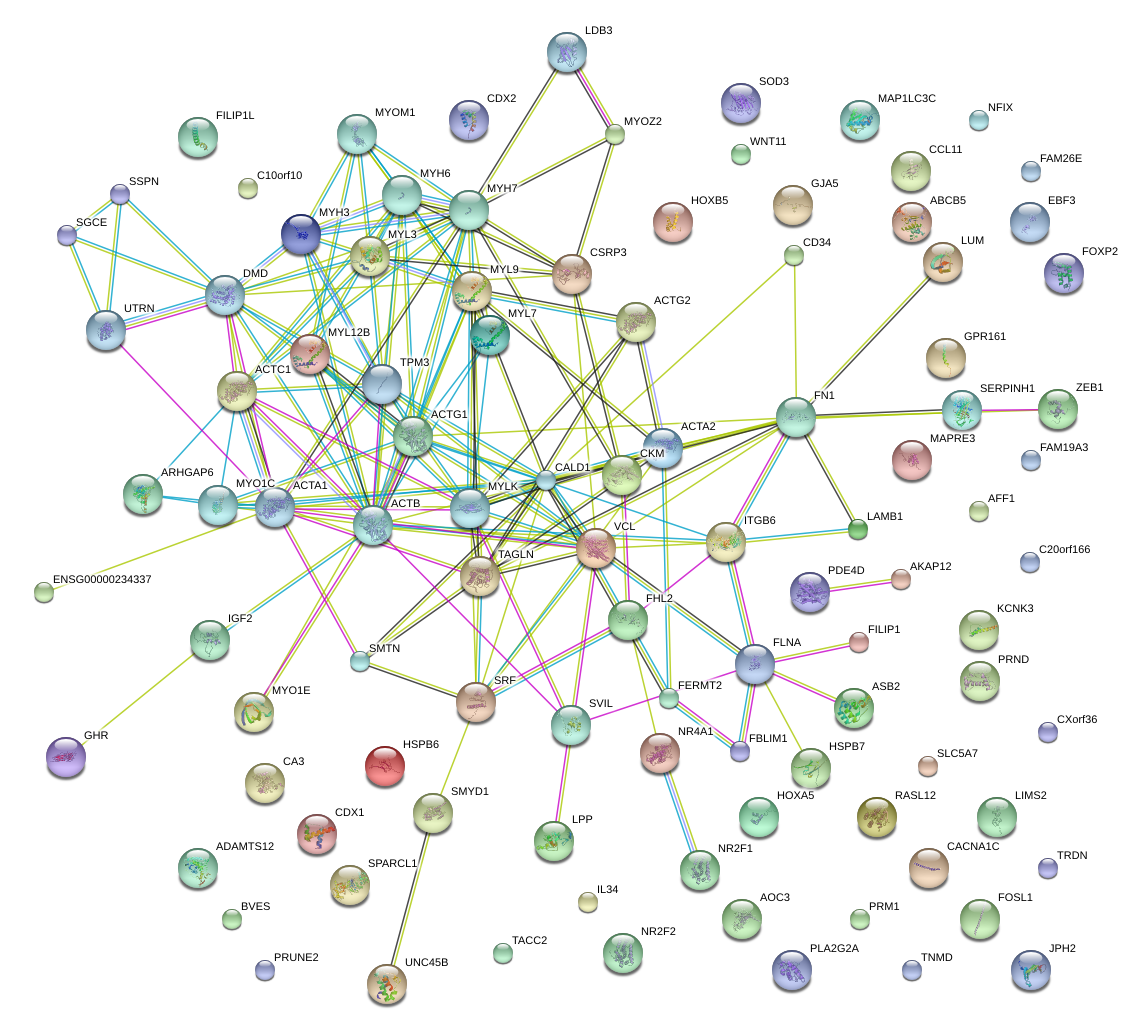
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr7_-_94285426
|
4.110
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr6_-_76203256
|
4.049
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr11_-_65667860
|
4.046
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr12_+_2162703
|
3.984
|
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chrX_-_11284094
|
3.977
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr6_+_43139190
|
3.944
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr12_-_91505541
|
3.878
|
NM_002345
|
LUM
|
lumican
|
|
chr20_+_61147659
|
3.864
|
NM_178463
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr10_+_123922940
|
3.850
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr10_+_31610063
|
3.829
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr9_-_79520878
|
3.769
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chrX_-_153602998
|
3.764
|
NM_001110556
NM_001456
|
FLNA
|
filamin A, alpha
|
|
chr10_-_45474210
|
3.695
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr11_-_19223516
|
3.668
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr17_-_79792923
|
3.644
|
NM_001007533
|
PPP1R27
|
protein phosphatase 1, regulatory subunit 27
|
|
chr7_+_134464161
|
3.612
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr11_-_75921785
|
3.492
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr1_-_168106810
|
3.484
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr11_-_2162167
|
3.456
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr7_-_44180911
|
3.423
|
NM_021223
|
MYL7
|
myosin, light chain 7, regulatory
|
|
chr3_+_187871473
|
3.401
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chrX_-_32430370
|
3.363
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr16_+_70657894
|
3.350
|
NM_001172772
|
IL34
|
interleukin 34
|
|
chr2_+_26915390
|
3.335
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr5_+_149546340
|
3.297
|
NM_001804
|
CDX1
|
caudal type homeobox 1
|
|
chr2_-_216248184
|
3.278
|
|
FN1
|
fibronectin 1
|
|
chr11_+_75273245
|
3.267
|
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr4_+_120056938
|
3.225
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr6_+_43139201
|
3.204
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr3_-_123339423
|
3.186
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr5_-_58295758
|
3.171
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_87928083
|
3.156
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr2_+_108602978
|
3.004
|
NM_021815
|
SLC5A7
|
solute carrier family 5 (choline transporter), member 7
|
|
chrX_-_45060081
|
2.985
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr17_-_46671043
|
2.969
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr1_-_147245483
|
2.966
|
NM_005266
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr1_+_113263188
|
2.926
|
NM_001004440
NM_182759
|
FAM19A3
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3
|
|
chr19_+_35849487
|
2.890
|
NM_005304
|
FFAR3
|
free fatty acid receptor 3
|
|
chr16_+_31483066
|
2.872
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr2_+_101179395
|
2.857
|
NM_024065
|
PDCL3
|
phosducin-like 3
|
|
chr14_+_63671101
|
2.830
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr4_+_87968191
|
2.801
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr15_+_33603162
|
2.791
|
NM_001036
NM_001243996
|
RYR3
|
ryanodine receptor 3
|
|
chr19_+_49376488
|
2.788
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr17_-_39684585
|
2.731
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr6_-_29527477
|
2.716
|
|
UBD
|
ubiquitin D
|
|
chr2_-_151344145
|
2.699
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chrX_-_10851808
|
2.660
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr10_+_6186758
|
2.627
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr11_-_123065836
|
2.618
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr12_-_50616425
|
2.593
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr15_+_45406518
|
2.564
|
NM_207581
|
DUOXA2
|
dual oxidase maturation factor 2
|
|
chr15_+_40643225
|
2.562
|
NM_001145643
|
PHGR1
|
proline/histidine/glycine-rich 1
|
|
chr19_+_16178170
|
2.481
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr3_-_123339178
|
2.414
|
|
MYLK
|
myosin light chain kinase
|
|
chr20_-_62680880
|
2.391
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr1_-_31845818
|
2.343
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr20_+_207898
|
2.271
|
NM_080831
|
DEFB129
|
defensin, beta 129
|
|
chr7_+_115863004
|
2.248
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr6_-_29527701
|
2.233
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr16_+_30077120
|
2.222
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr4_+_15376174
|
2.199
|
NM_001135171
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr17_+_41003360
|
2.199
|
|
|
|
|
chr21_-_47704166
|
2.162
|
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein
|
|
chr15_+_74218962
|
2.153
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr19_-_46283763
|
2.151
|
NM_001081563
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr20_+_33146500
|
2.141
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr16_+_30077146
|
2.127
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr10_-_16859408
|
2.107
|
NM_012425
NM_152724
|
RSU1
|
Ras suppressor protein 1
|
|
chr19_-_42894419
|
2.088
|
NM_032488
|
CNFN
|
cornifelin
|
|
chr7_-_559479
|
2.054
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr5_-_169816637
|
2.012
|
NM_004137
|
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1
|
|
chr10_-_16859389
|
1.993
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr19_+_54372659
|
1.971
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr2_-_96810775
|
1.963
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chrX_+_70521627
|
1.942
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr16_+_30076993
|
1.928
|
NM_001243177
NM_184041
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr8_-_22550708
|
1.910
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr9_+_139249251
|
1.906
|
NM_001200003
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr17_-_1389013
|
1.902
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr12_+_54426831
|
1.886
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chrX_-_153599719
|
1.884
|
|
FLNA
|
filamin A, alpha
|
|
chr15_-_45406345
|
1.796
|
NM_014080
|
DUOX2
|
dual oxidase 2
|
|
chr3_+_63953419
|
1.770
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr5_+_169816490
|
1.769
|
|
|
|
|
chr11_-_4718948
|
1.758
|
NM_030774
|
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2
|
|
chr7_-_80548322
|
1.720
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr7_-_80548666
|
1.716
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr7_+_114055234
|
1.713
|
|
FOXP2
|
forkhead box P2
|
|
chr4_-_76957139
|
1.668
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr6_+_144612872
|
1.589
|
NM_007124
|
UTRN
|
utrophin
|
|
chr10_+_64280206
|
1.581
|
NM_199452
|
ZNF365
|
zinc finger protein 365
|
|
chr18_+_61144143
|
1.567
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr12_+_50497790
|
1.553
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr1_+_181057835
|
1.540
|
|
IER5
|
immediate early response 5
|
|
chr1_+_43148048
|
1.515
|
NM_004559
|
YBX1
|
Y box binding protein 1
|
|
chr17_-_10372875
|
1.505
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr17_-_41132019
|
1.497
|
NM_025267
|
AARSD1
|
alanyl-tRNA synthetase domain containing 1
|
|
chr17_-_73505914
|
1.494
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr17_-_46671322
|
1.451
|
|
HOXB5
|
homeobox B5
|
|
chr7_+_114055171
|
1.450
|
|
FOXP2
|
forkhead box P2
|
|
chr19_+_1026582
|
1.435
|
|
CNN2
|
calponin 2
|
|
chr3_-_101405400
|
1.428
|
NM_000986
|
RPL24
|
ribosomal protein L24
|
|
chr13_-_78549663
|
1.415
|
NM_000115
|
EDNRB
|
endothelin receptor type B
|
|
chr6_-_29644930
|
1.409
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr5_-_124080773
|
1.391
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr10_+_6186903
|
1.376
|
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr4_+_186064416
|
1.372
|
NM_001151
|
SLC25A4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4
|
|
chr11_-_65667858
|
1.353
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr6_-_86388419
|
1.309
|
|
SNHG5
|
small nucleolar RNA host gene 5 (non-protein coding)
|
|
chr15_-_45406322
|
1.297
|
|
DUOX2
|
dual oxidase 2
|
|
chr1_-_935469
|
1.275
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr17_+_3379295
|
1.237
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr1_-_89591685
|
1.208
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr22_+_38004676
|
1.205
|
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr10_+_6186878
|
1.182
|
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr22_+_38004865
|
1.159
|
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr20_+_61147924
|
1.144
|
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr1_+_19970674
|
1.129
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|