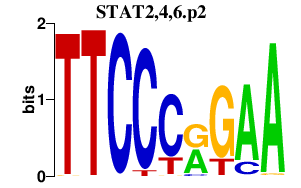
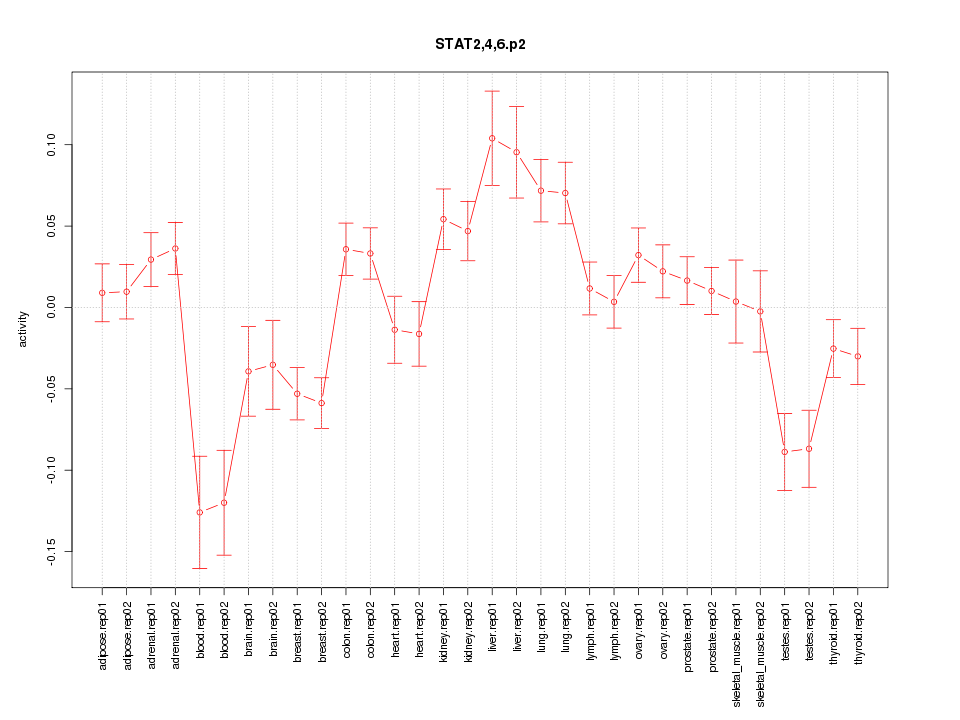
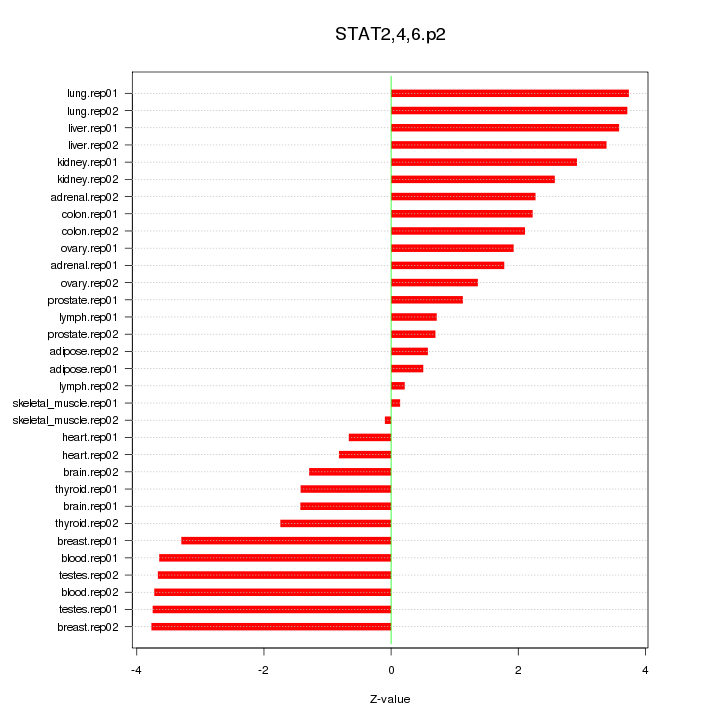
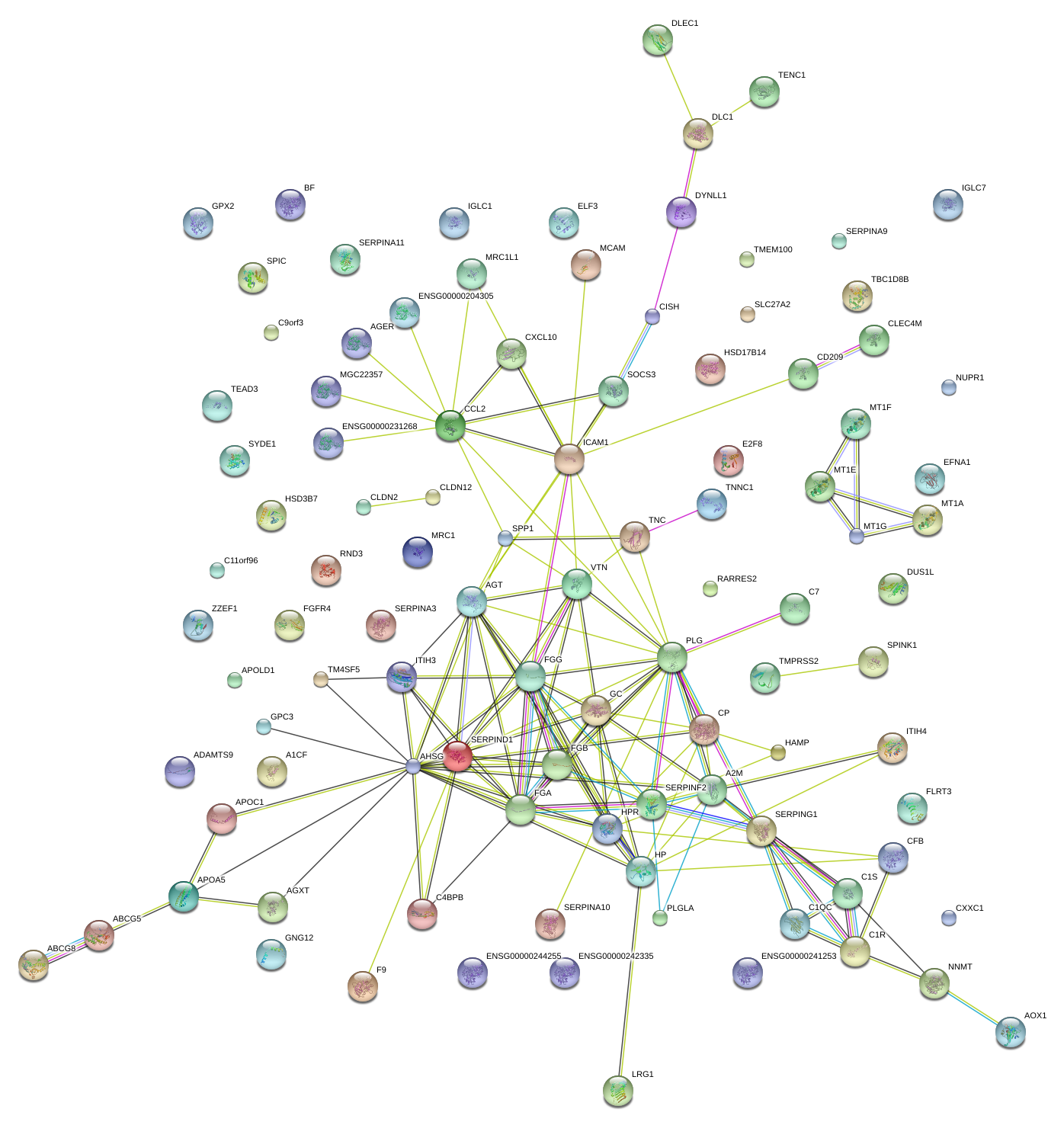
|
chr14_+_95078713
|
10.080
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr12_-_7245006
|
9.441
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr12_-_7245048
|
8.003
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr5_-_147211235
|
5.917
|
NM_003122
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr5_-_147211141
|
5.899
|
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr17_-_76356153
|
5.536
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr4_-_155533800
|
5.524
|
|
FGG
|
fibrinogen gamma chain
|
|
chr5_+_40909558
|
5.522
|
NM_000587
|
C7
|
complement component 7
|
|
chr12_+_53443834
|
5.487
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr4_-_155511838
|
5.421
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr4_-_155533882
|
5.324
|
NM_000509
NM_021870
|
FGG
|
fibrinogen gamma chain
|
|
chr12_-_9268522
|
5.320
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr4_-_155533757
|
5.199
|
|
FGG
|
fibrinogen gamma chain
|
|
chr5_+_40909617
|
5.072
|
|
C7
|
complement component 7
|
|
chr4_-_155533850
|
4.965
|
|
FGG
|
fibrinogen gamma chain
|
|
chr11_+_57365026
|
4.928
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr2_+_201450730
|
4.872
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr3_+_52828783
|
4.795
|
NM_002217
|
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3
|
|
chr17_+_1646129
|
4.769
|
NM_000934
NM_001165921
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr17_+_1646290
|
4.661
|
NM_001165920
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr5_-_121814781
|
4.592
|
|
|
|
|
chr1_+_201979689
|
4.585
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr1_-_68299047
|
4.562
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr5_+_40909487
|
4.505
|
|
C7
|
complement component 7
|
|
chr3_-_52864687
|
4.478
|
NM_001166449
NM_002218
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4
|
|
chr17_+_1646143
|
4.389
|
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr11_+_114167197
|
4.326
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr12_+_7167979
|
4.280
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr14_-_94919085
|
4.275
|
NM_001080451
|
SERPINA11
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11
|
|
chr1_+_22970117
|
4.191
|
NM_001114101
NM_172369
|
C1QC
|
complement component 1, q subcomponent, C chain
|
|
chr16_-_56701904
|
4.162
|
NM_005950
|
MT1G
|
metallothionein 1G
|
|
chr14_-_65409445
|
4.114
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr11_+_57365577
|
3.987
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr11_+_114167168
|
3.960
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr19_+_35773409
|
3.717
|
NM_021175
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chrX_+_106171328
|
3.715
|
|
CLDN2
|
claudin 2
|
|
chr19_+_15218141
|
3.690
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr2_+_201450579
|
3.612
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr11_-_119187811
|
3.575
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chrX_+_106045880
|
3.511
|
NM_017752
NM_198881
|
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain)
|
|
chr6_-_35464716
|
3.472
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr11_-_116663106
|
3.453
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr10_+_17851342
|
3.419
|
NM_002438
|
MRC1
|
mannose receptor, C type 1
|
|
chr12_-_9268440
|
3.315
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_-_230849863
|
3.275
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr16_+_72088507
|
3.258
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr1_+_201979747
|
3.247
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr7_-_150038689
|
3.229
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr12_-_9268475
|
3.218
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr3_+_186330844
|
3.206
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr19_+_45418063
|
3.178
|
|
APOC1
|
apolipoprotein C-I
|
|
chr3_+_186330859
|
3.144
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr12_-_9268513
|
3.125
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr4_+_155484131
|
3.080
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr6_+_31913852
|
3.052
|
|
CFB
|
complement factor B
|
|
chr3_-_50649110
|
3.043
|
NM_013324
NM_145071
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr16_-_28550227
|
3.041
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr5_+_176516550
|
3.040
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr17_+_4675175
|
3.031
|
NM_003963
|
TM4SF5
|
transmembrane 4 L six family member 5
|
|
chr3_-_148939784
|
3.028
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr6_+_31913920
|
3.000
|
|
CFB
|
complement factor B
|
|
chr2_-_151344145
|
2.978
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr16_+_30996462
|
2.929
|
NM_001142777
NM_001142778
NM_025193
|
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr1_+_207262583
|
2.884
|
NM_000716
NM_001017364
NM_001017367
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr3_+_186330711
|
2.834
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr1_-_230850335
|
2.828
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr1_-_230849847
|
2.796
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr4_+_88896838
|
2.780
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr7_+_90032771
|
2.769
|
|
CLDN12
|
claudin 12
|
|
chr19_-_4540035
|
2.750
|
NM_052972
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr15_+_50474357
|
2.721
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr6_+_31913763
|
2.720
|
|
CFB
|
complement factor B
|
|
chr1_+_155099935
|
2.692
|
|
EFNA1
|
ephrin-A1
|
|
chr17_+_32582295
|
2.683
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chrX_+_106143393
|
2.682
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr16_-_28550324
|
2.681
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr7_+_90032647
|
2.650
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr4_-_72671236
|
2.647
|
NM_001204306
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chrX_+_138612894
|
2.638
|
NM_000133
|
F9
|
coagulation factor IX
|
|
chr19_+_7828034
|
2.636
|
NM_001144904
NM_001144905
NM_001144906
NM_001144907
NM_001144908
NM_001144909
NM_001144910
NM_001144911
NM_014257
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr19_-_49339667
|
2.618
|
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr6_+_31913708
|
2.600
|
NM_001710
|
CFB
|
complement factor B
|
|
chr4_+_88896801
|
2.598
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr14_-_94759594
|
2.543
|
NM_001100607
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10
|
|
chr21_-_42879908
|
2.533
|
NM_001135099
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr4_+_88896868
|
2.509
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr10_-_52645430
|
2.505
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr11_+_43963809
|
2.504
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr9_-_117880411
|
2.456
|
NM_002160
|
TNC
|
tenascin C
|
|
chr17_-_53800074
|
2.449
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr17_-_26697358
|
2.433
|
NM_000638
|
VTN
|
vitronectin
|
|
chr20_-_14318200
|
2.421
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr4_+_22694547
|
2.408
|
NM_001128432
NM_020973
|
GBA3
|
glucosidase, beta, acid 3 (cytosolic)
|
|
chr17_-_26697224
|
2.404
|
|
VTN
|
vitronectin
|
|
chr19_+_10381516
|
2.397
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr1_-_230849936
|
2.389
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr19_+_10381763
|
2.369
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr3_-_64673330
|
2.368
|
NM_182920
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr12_+_12938540
|
2.341
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr17_-_26697310
|
2.333
|
|
VTN
|
vitronectin
|
|
chr6_-_32151996
|
2.318
|
NM_001136
NM_001206929
NM_001206932
NM_001206934
NM_001206936
NM_001206940
NM_001206954
NM_001206966
NM_172197
|
AGER
|
advanced glycosylation end product-specific receptor
|
|
chr4_+_88896899
|
2.314
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_-_44065888
|
2.303
|
NM_022436
|
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5
|
|
chrX_-_133119475
|
2.292
|
|
GPC3
|
glypican 3
|
|
chr2_+_44066102
|
2.286
|
NM_022437
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8
|
|
chr22_+_21128378
|
2.282
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr9_+_97766414
|
2.272
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr6_+_161123224
|
2.265
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr5_+_38846260
|
2.253
|
|
OSMR
|
oncostatin M receptor
|
|
chr1_+_209859524
|
2.236
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr1_+_32712817
|
2.229
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chr17_-_26697293
|
2.214
|
|
VTN
|
vitronectin
|
|
chr20_-_49308074
|
2.193
|
|
|
|
|
chr19_-_49339913
|
2.181
|
NM_016246
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr5_+_38845962
|
2.171
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr16_+_67204404
|
2.153
|
NM_001185058
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr5_-_138842291
|
2.153
|
NM_001077693
|
ECSCR
|
endothelial cell-specific chemotaxis regulator
|
|
chr4_-_101439071
|
2.144
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr3_-_158450230
|
2.140
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr11_+_58695101
|
2.127
|
NM_001220494
|
GLYATL1
|
glycine-N-acyltransferase-like 1
|
|
chr10_+_74653338
|
2.121
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr6_+_30130982
|
2.119
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr7_-_120498009
|
2.115
|
|
TSPAN12
|
tetraspanin 12
|
|
chr11_-_6462129
|
2.101
|
NM_000613
|
HPX
|
hemopexin
|
|
chr15_+_84322809
|
2.089
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr3_-_158450271
|
2.087
|
NM_002888
NM_206963
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr15_-_45493372
|
2.080
|
NM_138356
|
SHF
|
Src homology 2 domain containing F
|
|
chrX_+_73641084
|
2.072
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chrX_+_152770083
|
2.049
|
|
BGN
|
biglycan
|
|
chr17_+_32683470
|
2.046
|
NM_005408
|
CCL13
|
chemokine (C-C motif) ligand 13
|
|
chr7_+_134464417
|
2.038
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_27693336
|
2.032
|
NM_004672
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chr3_-_158450205
|
2.025
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr3_-_50649080
|
2.021
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr7_+_117120016
|
2.012
|
NM_000492
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7)
|
|
chr12_+_93964801
|
2.005
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chrX_-_133119624
|
2.001
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr2_-_75796847
|
1.994
|
NM_032181
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr10_+_88728187
|
1.980
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr12_+_53442972
|
1.975
|
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr19_-_7812396
|
1.971
|
NM_001144893
NM_001144894
NM_001144895
NM_001144896
NM_001144897
NM_001144899
NM_021155
|
CD209
|
CD209 molecule
|
|
chr5_+_57878901
|
1.962
|
NM_138453
|
RAB3C
|
RAB3C, member RAS oncogene family
|
|
chr3_-_124653571
|
1.942
|
NM_033049
|
MUC13
|
mucin 13, cell surface associated
|
|
chr5_+_71502700
|
1.911
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr14_-_53417272
|
1.884
|
|
FERMT2
|
fermitin family member 2
|
|
chr3_+_171561138
|
1.880
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr7_-_120498128
|
1.877
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr1_+_207262211
|
1.857
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr22_+_38598065
|
1.849
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr16_-_20338808
|
1.841
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr1_-_11107150
|
1.838
|
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr2_+_234959345
|
1.822
|
NM_006944
|
SPP2
|
secreted phosphoprotein 2, 24kDa
|
|
chr6_+_31895401
|
1.815
|
|
C2
|
complement component 2
|
|
chr2_+_108994366
|
1.801
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr10_+_75673346
|
1.792
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr3_-_184095927
|
1.790
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr6_-_46889620
|
1.790
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr12_-_93964636
|
1.783
|
|
|
|
|
chr19_-_44860798
|
1.778
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr10_-_52645384
|
1.774
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr14_-_54423448
|
1.769
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr19_+_35532617
|
1.752
|
|
HPN
|
hepsin
|
|
chr19_-_5839702
|
1.731
|
NM_000150
NM_001040701
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr20_+_36974823
|
1.725
|
NM_004139
|
LBP
|
lipopolysaccharide binding protein
|
|
chr5_-_102898450
|
1.724
|
NM_031438
|
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12
|
|
chr8_+_18248754
|
1.696
|
NM_000015
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase)
|
|
chr4_+_77356252
|
1.691
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr1_-_204120909
|
1.687
|
|
ETNK2
|
ethanolamine kinase 2
|
|
chr5_+_42549657
|
1.681
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr17_-_73505914
|
1.679
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr14_+_38678280
|
1.675
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr2_+_79347610
|
1.674
|
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr13_+_111134933
|
1.667
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr19_-_4540078
|
1.656
|
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr2_+_79347487
|
1.645
|
NM_002909
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr1_+_22138757
|
1.629
|
NM_001013693
|
LDLRAD2
|
low density lipoprotein receptor class A domain containing 2
|
|
chrX_-_99891690
|
1.625
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr16_-_52581713
|
1.616
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr4_-_186697065
|
1.607
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_101841593
|
1.605
|
NM_001308
|
CPN1
|
carboxypeptidase N, polypeptide 1
|
|
chr2_+_233498206
|
1.601
|
NM_025202
|
EFHD1
|
EF-hand domain family, member D1
|
|
chr7_+_134464372
|
1.599
|
|
CALD1
|
caldesmon 1
|
|
chr4_+_174451608
|
1.583
|
|
NBLA00301
|
Nbla00301
|
|
chr2_-_31805968
|
1.580
|
NM_000348
|
SRD5A2
|
steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2)
|
|
chr3_+_52812438
|
1.578
|
NM_001166434
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr15_+_33010174
|
1.572
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr1_+_46640748
|
1.570
|
NM_005727
|
TSPAN1
|
tetraspanin 1
|
|
chr7_-_100239136
|
1.570
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr16_-_75285383
|
1.551
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr2_-_190044330
|
1.548
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr12_+_52627007
|
1.547
|
|
KRT7
|
keratin 7
|
|
chrX_+_37208527
|
1.540
|
NM_000950
NM_001142395
NM_001173486
NM_001173489
NM_001173490
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1
|
|
chr10_-_90750955
|
1.529
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr6_-_31845446
|
1.524
|
NM_001178045
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr1_+_95285900
|
1.519
|
NM_001114106
|
SLC44A3
|
solute carrier family 44, member 3
|
|
chr19_-_36001336
|
1.516
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr6_-_28304151
|
1.508
|
NM_001243241
NM_001243242
NM_001243244
NM_030899
|
ZNF323
|
zinc finger protein 323
|
|
chr1_-_201368716
|
1.500
|
|
LAD1
|
ladinin 1
|
|
chr20_-_45313122
|
1.496
|
NM_001011554
NM_001193340
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr2_+_27301434
|
1.494
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|