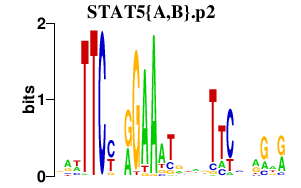
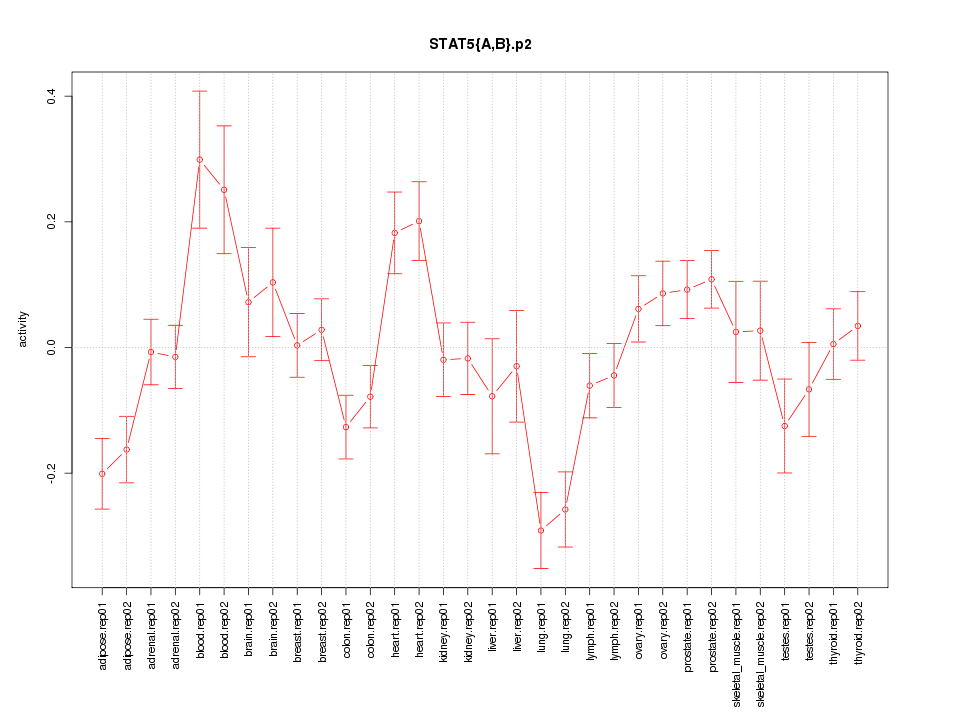
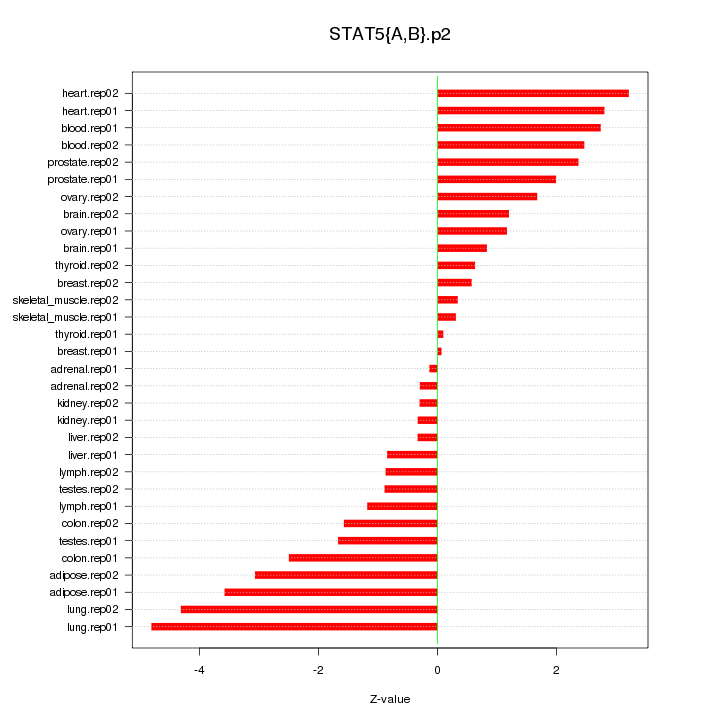
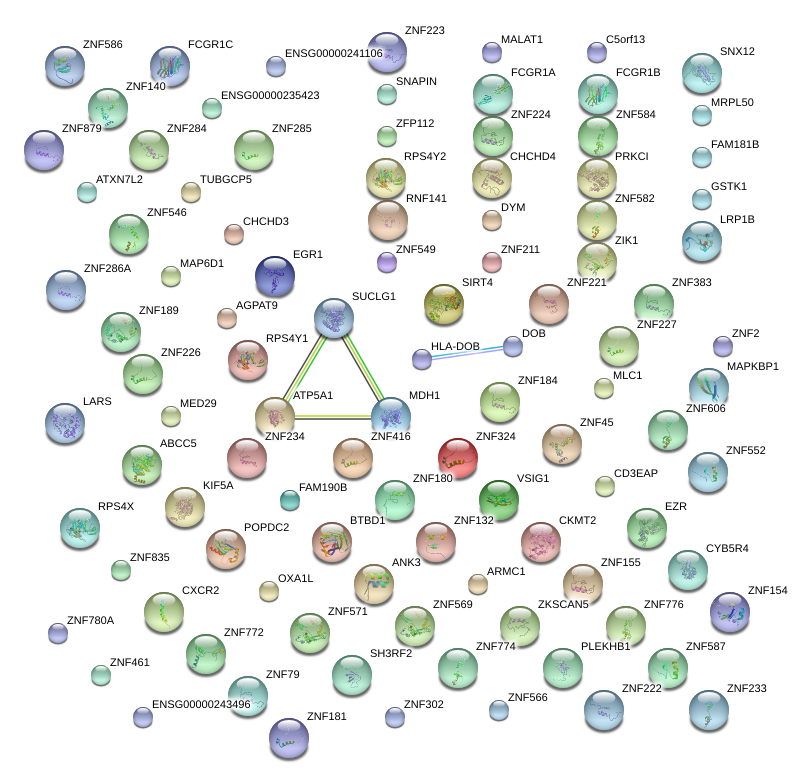
|
chr19_+_44556163
|
5.450
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chrX_+_107288199
|
3.825
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr19_+_37709068
|
3.439
|
|
ZNF383
|
zinc finger protein 383
|
|
chr19_+_35225124
|
3.260
|
|
ZNF181
|
zinc finger protein 181
|
|
chr1_+_149754245
|
3.195
|
NM_000566
|
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64)
|
|
chr1_-_120935893
|
3.186
|
|
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64)
|
|
chr1_-_120935943
|
3.180
|
NM_001004340
NM_001017986
NM_001244910
|
FCGR1A
FCGR1B
|
Fc fragment of IgG, high affinity Ia, receptor (CD64)
Fc fragment of IgG, high affinity Ib, receptor (CD64)
|
|
chr9_+_130186652
|
3.038
|
NM_007135
|
ZNF79
|
zinc finger protein 79
|
|
chr19_-_57183122
|
2.949
|
NM_001005850
|
ZNF835
|
zinc finger protein 835
|
|
chr19_-_57988891
|
2.920
|
NM_001024596
NM_001144068
|
ZNF772
|
zinc finger protein 772
|
|
chr19_+_44331443
|
2.822
|
NM_181845
|
ZNF283
|
zinc finger protein 283
|
|
chr19_+_35225095
|
2.770
|
|
ZNF181
|
zinc finger protein 181
|
|
chr2_+_63816092
|
2.746
|
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr18_-_43684198
|
2.661
|
NM_001001937
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr15_+_90895476
|
2.594
|
NM_001004309
|
ZNF774
|
zinc finger protein 774
|
|
chr19_-_40596781
|
2.586
|
NM_001010880
NM_001142577
NM_001142578
NM_001142579
|
ZNF780A
|
zinc finger protein 780A
|
|
chr3_-_183735617
|
2.557
|
NM_001023587
NM_005688
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr19_+_44507076
|
2.484
|
NM_006300
|
ZNF230
|
zinc finger protein 230
|
|
chr19_+_40502942
|
2.481
|
NM_178544
|
ZNF546
|
zinc finger protein 546
|
|
chr19_-_44439410
|
2.476
|
NM_003425
|
ZNF45
|
zinc finger protein 45
|
|
chrY_+_2709596
|
2.413
|
NM_001008
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr19_+_44455379
|
2.407
|
NM_013359
|
ZNF221
|
zinc finger protein 221
|
|
chr2_+_95831533
|
2.383
|
|
ZNF2
|
zinc finger protein 2
|
|
chr19_+_44617510
|
2.369
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr19_-_45004482
|
2.352
|
NM_013256
|
ZNF180
|
zinc finger protein 180
|
|
chr19_+_58038744
|
2.341
|
|
ZNF549
|
zinc finger protein 549
|
|
chr19_-_44405934
|
2.336
|
|
LOC100505715
|
uncharacterized LOC100505715
|
|
chr19_+_44764032
|
2.308
|
NM_001207005
NM_181756
|
ZNF233
|
zinc finger protein 233
|
|
chrY_+_2709630
|
2.295
|
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr2_+_63816143
|
2.233
|
NM_001199111
NM_001199112
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr19_+_44576266
|
2.188
|
NM_001037813
|
ZNF284
|
zinc finger protein 284
|
|
chr19_-_56904869
|
2.152
|
NM_144690
|
ZNF582
|
zinc finger protein 582
|
|
chr19_+_58258160
|
2.134
|
NM_173632
|
ZNF776
|
zinc finger protein 776
|
|
chr5_+_145317297
|
2.126
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr9_+_130186714
|
2.125
|
|
ZNF79
|
zinc finger protein 79
|
|
chr19_+_44529493
|
2.102
|
NM_001129996
NM_013360
|
ZNF284
ZNF222
|
zinc finger protein 284
zinc finger protein 222
|
|
chr19_+_58038692
|
2.091
|
NM_001199295
NM_153263
|
ZNF549
|
zinc finger protein 549
|
|
chr19_+_35168558
|
2.083
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr11_+_73357222
|
1.976
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr19_-_58326280
|
1.934
|
NM_024762
|
ZNF552
|
zinc finger protein 552
|
|
chr2_+_95831176
|
1.909
|
NM_001017396
NM_021088
|
ZNF2
|
zinc finger protein 2
|
|
chr19_+_58258189
|
1.907
|
|
ZNF776
|
zinc finger protein 776
|
|
chr1_+_153631144
|
1.860
|
NM_012437
|
SNAPIN
|
SNAP-associated protein
|
|
chr1_+_153631193
|
1.834
|
|
SNAPIN
|
SNAP-associated protein
|
|
chr9_+_104161118
|
1.804
|
NM_003452
NM_197977
|
ZNF189
|
zinc finger protein 189
|
|
chr4_+_84457256
|
1.798
|
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr8_-_66546394
|
1.789
|
|
ARMC1
|
armadillo repeat containing 1
|
|
chr19_-_58090071
|
1.777
|
NM_017879
|
ZNF416
|
zinc finger protein 416
|
|
chr19_+_35225479
|
1.768
|
NM_001029997
NM_001145665
|
ZNF181
|
zinc finger protein 181
|
|
chr19_-_37958338
|
1.724
|
NM_152484
|
ZNF569
|
zinc finger protein 569
|
|
chr22_-_50523853
|
1.714
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr6_-_27440459
|
1.696
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr19_+_58144510
|
1.666
|
NM_006385
NM_198855
|
ZNF211
|
zinc finger protein 211
|
|
chr8_-_66546400
|
1.650
|
|
ARMC1
|
armadillo repeat containing 1
|
|
chr3_-_183543295
|
1.616
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr19_+_44488331
|
1.614
|
NM_003445
NM_198089
|
ZNF155
|
zinc finger protein 155
|
|
chr19_+_58281049
|
1.579
|
|
ZNF586
|
zinc finger protein 586
|
|
chr10_+_86184685
|
1.569
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr5_-_111093251
|
1.524
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr22_-_50523670
|
1.468
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr12_+_120740123
|
1.458
|
NM_012240
|
SIRT4
|
sirtuin 4
|
|
chr3_-_14166370
|
1.453
|
NM_001098502
NM_144636
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr19_+_44598479
|
1.443
|
NM_013398
|
ZNF224
|
zinc finger protein 224
|
|
chr19_-_58090254
|
1.408
|
|
ZNF416
|
zinc finger protein 416
|
|
chr3_-_14166228
|
1.402
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr19_+_58095507
|
1.398
|
NM_001010879
|
ZIK1
|
zinc finger protein interacting with K protein 1 homolog (mouse)
|
|
chr15_+_22833394
|
1.391
|
NM_001102610
NM_052903
|
TUBGCP5
|
tubulin, gamma complex associated protein 5
|
|
chr3_-_14166292
|
1.387
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr19_+_44645708
|
1.383
|
NM_001144824
NM_006630
|
ZNF234
|
zinc finger protein 234
|
|
chr7_+_99102272
|
1.382
|
NM_145102
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr19_+_58920118
|
1.349
|
|
ZNF584
|
zinc finger protein 584
|
|
chr19_+_39882018
|
1.261
|
|
MED29
|
mediator complex subunit 29
|
|
chr15_-_83735998
|
1.259
|
|
BTBD1
|
BTB (POZ) domain containing 1
|
|
chr19_-_44905754
|
1.257
|
NM_152354
|
ZNF285
ZFP112
|
zinc finger protein 285
zinc finger protein 112 homolog (mouse)
|
|
chr19_-_58951588
|
1.251
|
NM_003433
|
ZNF132
|
zinc finger protein 132
|
|
chr11_+_65268313
|
1.246
|
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr7_+_142960588
|
1.242
|
|
GSTK1
|
glutathione S-transferase kappa 1
|
|
chr19_-_58220535
|
1.240
|
NM_001085384
|
ZNF154
|
zinc finger protein 154
|
|
chr19_+_58280996
|
1.178
|
NM_001077426
NM_001204814
NM_017652
|
ZNF586
|
zinc finger protein 586
|
|
chr3_-_119379269
|
1.177
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr9_-_104160876
|
1.167
|
NM_019051
|
MRPL50
|
mitochondrial ribosomal protein L50
|
|
chr7_-_112758604
|
1.167
|
|
LOC401397
|
uncharacterized LOC401397
|
|
chr7_-_132766834
|
1.157
|
|
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3
|
|
chr15_+_42066690
|
1.152
|
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr19_-_36980336
|
1.149
|
NM_032838
NM_001145343
NM_001145344
NM_001145345
|
ZNF566
|
zinc finger protein 566
|
|
chr18_-_46987016
|
1.142
|
NM_017653
|
DYM
|
dymeclin
|
|
chr14_+_23235730
|
1.140
|
NM_005015
|
OXA1L
|
oxidase (cytochrome c) assembly 1-like
|
|
chr19_+_58920007
|
1.128
|
NM_173548
|
ZNF584
|
zinc finger protein 584
|
|
chr3_-_14166246
|
1.116
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr8_-_66546413
|
1.111
|
NM_018120
|
ARMC1
|
armadillo repeat containing 1
|
|
chr12_+_133657478
|
1.098
|
|
ZNF140
|
zinc finger protein 140
|
|
chr9_-_115774471
|
1.092
|
NM_001101338
|
ZNF883
|
zinc finger protein 883
|
|
chr19_+_44645728
|
1.082
|
|
ZNF234
|
zinc finger protein 234
|
|
chr19_-_36980758
|
1.068
|
|
ZNF566
|
zinc finger protein 566
|
|
chr1_+_54519259
|
1.067
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr5_-_145562068
|
1.035
|
|
LARS
|
leucyl-tRNA synthetase
|
|
chr6_-_32784727
|
1.025
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr2_-_142888540
|
1.019
|
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr19_-_58514136
|
1.000
|
|
ZNF606
|
zinc finger protein 606
|
|
chr10_-_62493077
|
0.987
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_+_44716683
|
0.972
|
NM_182490
|
ZNF227
|
zinc finger protein 227
|
|
chr1_+_110026452
|
0.963
|
NM_153340
|
ATXN7L2
|
ataxin 7-like 2
|
|
chr19_+_51152634
|
0.959
|
NM_001195076
|
C19orf81
|
chromosome 19 open reading frame 81
|
|
chr19_-_37157718
|
0.950
|
NM_153257
|
ZNF461
|
zinc finger protein 461
|
|
chr6_+_84569394
|
0.946
|
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr6_-_159240398
|
0.939
|
|
EZR
|
ezrin
|
|
chr7_-_112758611
|
0.938
|
|
LOC401397
|
uncharacterized LOC401397
|
|
chr19_+_45909868
|
0.930
|
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr3_+_169940246
|
0.922
|
|
PRKCI
|
protein kinase C, iota
|
|
chr7_+_99102601
|
0.921
|
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr17_+_15603046
|
0.918
|
|
ZNF286A
|
zinc finger protein 286A
|
|
chr2_-_84686585
|
0.918
|
NM_003849
|
SUCLG1
|
succinate-CoA ligase, alpha subunit
|
|
chr6_+_84569427
|
0.916
|
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr22_-_50523780
|
0.914
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr5_+_178450775
|
0.912
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr5_+_80529138
|
0.898
|
NM_001099735
NM_001099736
NM_001825
|
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric)
|
|
chr7_+_99102572
|
0.894
|
NM_014569
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr2_-_63815761
|
0.891
|
NM_015910
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chr2_+_218989997
|
0.888
|
NM_001168298
|
CXCR2
|
chemokine (C-X-C motif) receptor 2
|
|
chr3_+_169940223
|
0.885
|
|
PRKCI
|
protein kinase C, iota
|
|
chr5_-_145562214
|
0.877
|
|
LARS
|
leucyl-tRNA synthetase
|
|
chr2_+_63815742
|
0.869
|
NM_005917
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr20_-_48530275
|
0.862
|
NM_001135773
|
SPATA2
|
spermatogenesis associated 2
|
|
chr19_-_37019169
|
0.845
|
NM_001012756
NM_001166036
NM_001166037
NM_001166038
|
ZNF260
|
zinc finger protein 260
|
|
chr19_+_58341661
|
0.841
|
NM_001204818
|
LOC100293516
|
zinc finger protein
|
|
chr4_+_2883636
|
0.831
|
|
ADD1
|
adducin 1 (alpha)
|
|
chr19_-_37341168
|
0.817
|
NM_001242801
|
ZNF790
|
zinc finger protein 790
|
|
chr9_+_137004913
|
0.817
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr6_-_159240367
|
0.805
|
|
EZR
|
ezrin
|
|
chr16_-_70472920
|
0.802
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr7_-_132766697
|
0.787
|
NM_017812
|
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3
|
|
chr6_-_159240399
|
0.787
|
|
EZR
|
ezrin
|
|
chr22_-_50523737
|
0.783
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr4_+_130017281
|
0.781
|
NM_001099783
|
C4orf33
|
chromosome 4 open reading frame 33
|
|
chr12_-_46384346
|
0.772
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr2_+_172290796
|
0.769
|
|
DCAF17
|
DDB1 and CUL4 associated factor 17
|
|
chr13_+_115079943
|
0.763
|
NM_001164144
NM_001164145
NM_032436
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr16_+_53468373
|
0.738
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr19_-_44860798
|
0.724
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr19_+_58331067
|
0.720
|
|
LOC100293516
|
zinc finger protein
|
|
chr8_-_99306563
|
0.707
|
NM_024759
|
NIPAL2
|
NIPA-like domain containing 2
|
|
chr17_-_48945338
|
0.705
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr3_-_151047309
|
0.702
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chrX_+_105412297
|
0.699
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr4_+_86396251
|
0.686
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr17_-_26697293
|
0.681
|
|
VTN
|
vitronectin
|
|
chr19_-_38183135
|
0.679
|
NM_152605
|
ZNF781
|
zinc finger protein 781
|
|
chr15_-_83735892
|
0.676
|
|
BTBD1
|
BTB (POZ) domain containing 1
|
|
chr19_-_38085646
|
0.673
|
NM_016536
|
ZNF571
|
zinc finger protein 571
|
|
chr6_+_84569364
|
0.670
|
NM_016230
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr19_+_9251052
|
0.669
|
NM_001190791
NM_020933
|
ZNF317
|
zinc finger protein 317
|
|
chr19_-_58427934
|
0.664
|
NM_152475
|
ZNF417
|
zinc finger protein 417
|
|
chr22_-_38902300
|
0.663
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr17_-_39296702
|
0.663
|
NM_030976
|
KRTAP4-6
|
keratin associated protein 4-6
|
|
chr6_-_116601085
|
0.659
|
|
TSPYL1
|
TSPY-like 1
|
|
chr17_-_3794036
|
0.656
|
NM_172206
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha
|
|
chr7_+_92861652
|
0.642
|
NM_017667
NM_024553
|
CCDC132
|
coiled-coil domain containing 132
|
|
chr15_-_83736105
|
0.635
|
NM_001011885
NM_025238
|
BTBD1
|
BTB (POZ) domain containing 1
|
|
chr18_+_71815745
|
0.629
|
NM_014177
|
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast)
|
|
chr16_+_53468362
|
0.627
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr12_-_133532847
|
0.627
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr3_+_196466806
|
0.624
|
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr19_+_17378264
|
0.616
|
|
BABAM1
|
BRISC and BRCA1 A complex member 1
|
|
chr2_-_63815581
|
0.613
|
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chr2_-_231084600
|
0.613
|
NM_004509
NM_004510
NM_080424
|
SP110
|
SP110 nuclear body protein
|
|
chr19_+_17416476
|
0.612
|
NM_023937
|
MRPL34
|
mitochondrial ribosomal protein L34
|
|
chr1_+_3773844
|
0.602
|
NM_004402
|
DFFB
|
DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase)
|
|
chr3_+_66119284
|
0.598
|
NM_173471
|
SLC25A26
|
solute carrier family 25, member 26
|
|
chr22_-_38902344
|
0.596
|
NM_001098504
NM_006386
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr19_-_37406918
|
0.596
|
NM_001171979
|
ZNF829
|
zinc finger protein 829
|
|
chr12_+_133657439
|
0.594
|
|
ZNF140
|
zinc finger protein 140
|
|
chr19_+_57946662
|
0.593
|
NM_001023561
|
ZNF749
|
zinc finger protein 749
|
|
chr7_+_110731061
|
0.593
|
NM_001099658
NM_001099660
NM_018334
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr7_+_5862822
|
0.588
|
|
|
|
|
chr19_-_58446739
|
0.588
|
NM_133460
|
ZNF418
|
zinc finger protein 418
|
|
chr12_-_46384208
|
0.585
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr11_+_134123406
|
0.583
|
NM_014384
|
ACAD8
|
acyl-CoA dehydrogenase family, member 8
|
|
chr17_-_26697310
|
0.582
|
|
VTN
|
vitronectin
|
|
chr5_+_85913762
|
0.581
|
NM_001867
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr19_+_39881950
|
0.579
|
NM_017592
|
MED29
|
mediator complex subunit 29
|
|
chr19_-_44809126
|
0.575
|
NM_004234
|
ZNF235
|
zinc finger protein 235
|
|
chr16_+_53468344
|
0.573
|
NM_005611
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr1_+_149871154
|
0.572
|
NM_016074
|
BOLA1
|
bolA homolog 1 (E. coli)
|
|
chr2_-_44223116
|
0.570
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr19_-_37341688
|
0.569
|
NM_001242800
|
ZNF790
|
zinc finger protein 790
|
|
chr19_+_58111244
|
0.565
|
NM_020880
|
ZNF530
|
zinc finger protein 530
|
|
chr20_-_43280359
|
0.563
|
NM_000022
|
ADA
|
adenosine deaminase
|
|
chr22_+_20748397
|
0.560
|
NM_003426
|
ZNF74
|
zinc finger protein 74
|
|
chr7_-_150754934
|
0.556
|
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr8_+_27950583
|
0.555
|
NM_018091
|
ELP3
|
elongation protein 3 homolog (S. cerevisiae)
|
|
chr19_+_17378148
|
0.546
|
NM_001033549
NM_014173
|
BABAM1
|
BRISC and BRCA1 A complex member 1
|
|
chr2_-_27886443
|
0.543
|
NM_014860
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like
|
|
chr17_-_39274587
|
0.542
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr16_+_53468406
|
0.531
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr6_+_116782555
|
0.529
|
NM_001010919
|
FAM26F
|
family with sequence similarity 26, member F
|
|
chr20_-_33680601
|
0.526
|
NM_015638
NM_199368
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr1_+_244217283
|
0.526
|
|
|
|
|
chr2_-_44223112
|
0.515
|
|
LRPPRC
|
leucine-rich PPR-motif containing
|
|
chr16_+_53468390
|
0.508
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr2_-_241500459
|
0.508
|
|
ANKMY1
|
ankyrin repeat and MYND domain containing 1
|