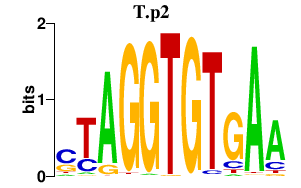
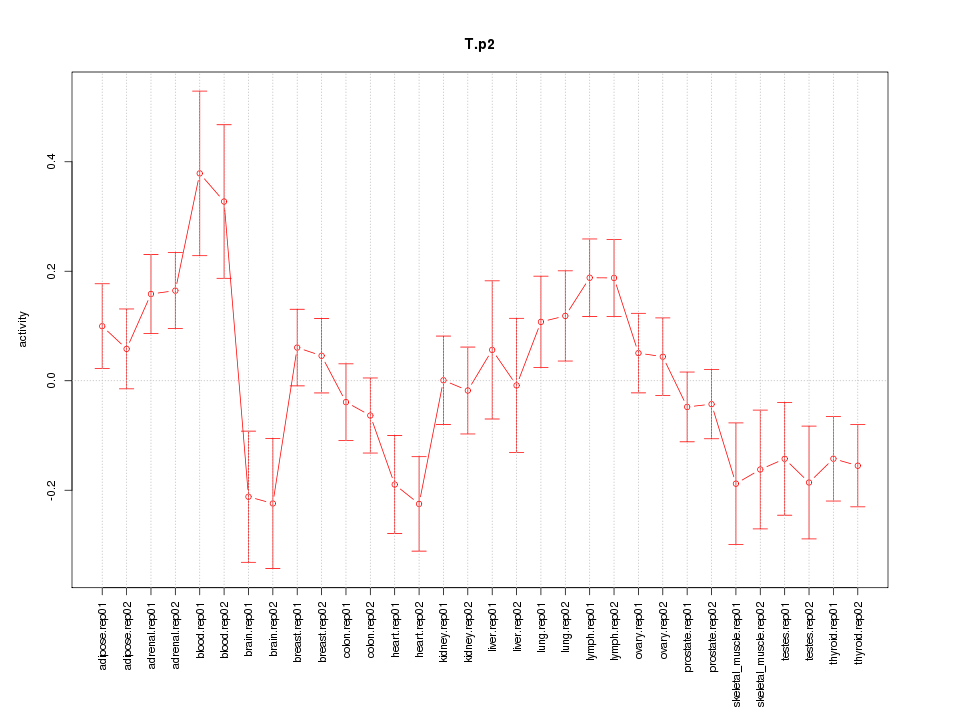
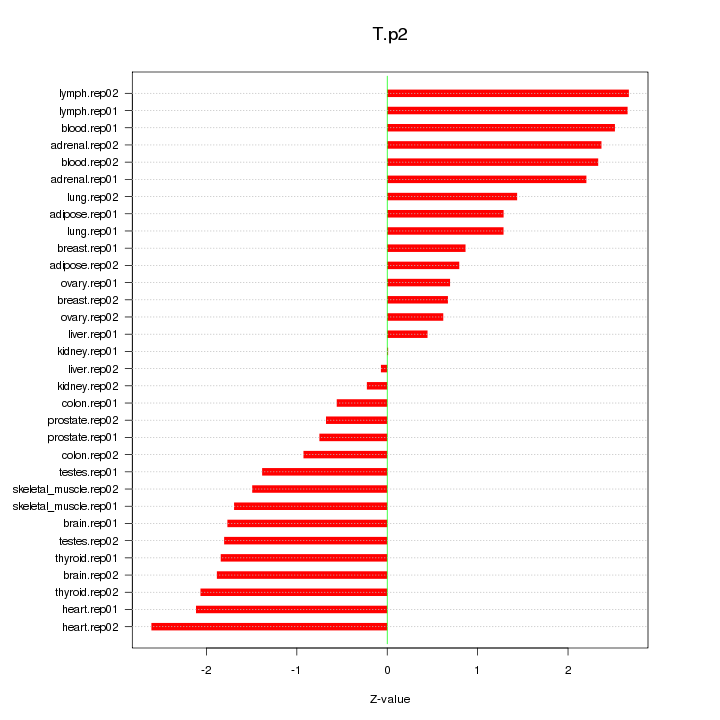
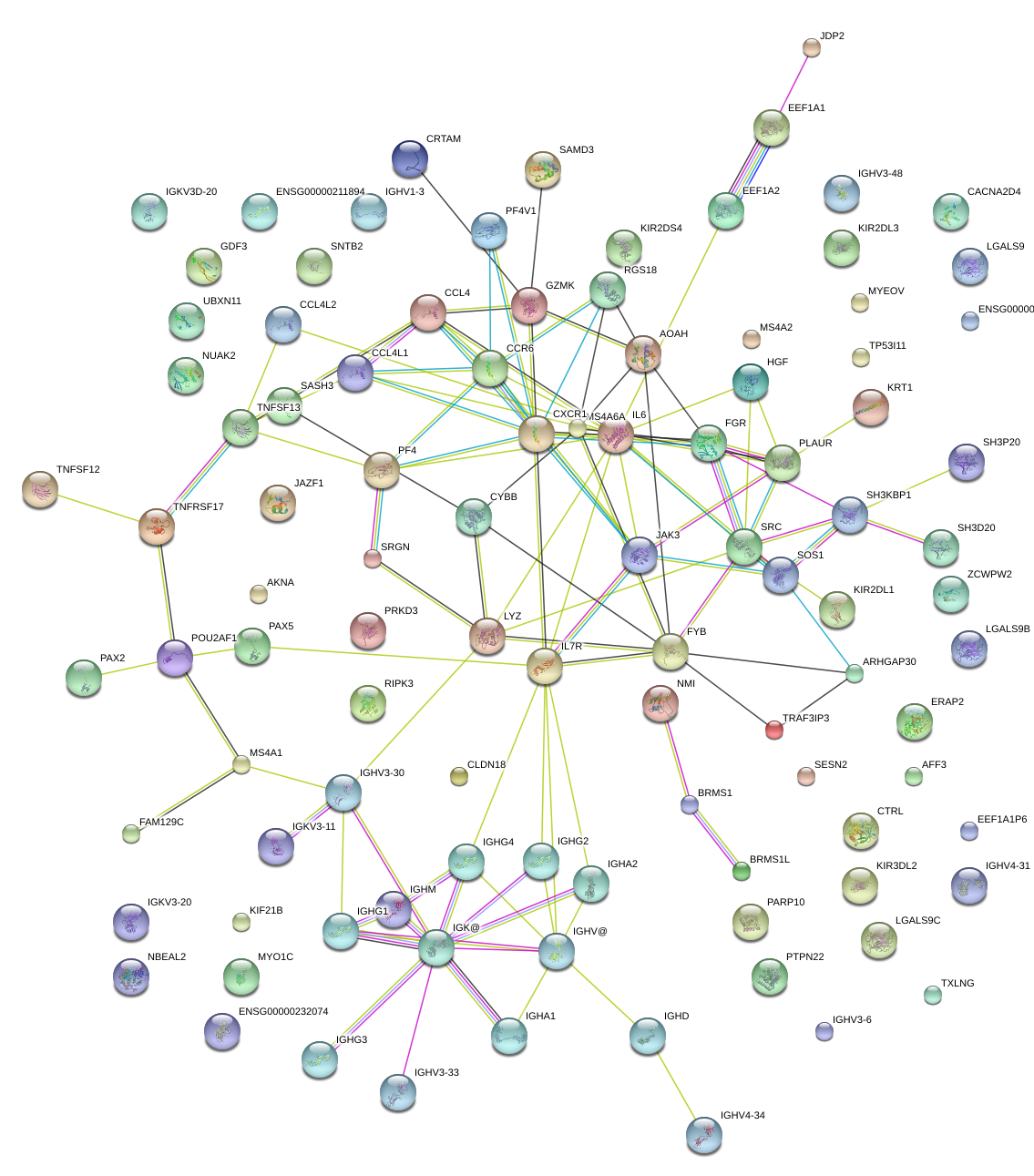
|
chr5_+_54320083
|
14.203
|
NM_002104
|
GZMK
|
granzyme K (granzyme 3; tryptase II)
|
|
chr14_-_107113943
|
11.666
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_+_128913881
|
10.445
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr14_-_107013474
|
10.375
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr6_-_130536417
|
9.761
|
NM_152552
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr4_-_74847669
|
8.953
|
NM_002619
|
PF4
|
platelet factor 4
|
|
chrX_+_128913897
|
8.729
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr17_+_34431219
|
8.376
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr14_-_107211131
|
7.826
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_+_209941894
|
7.709
|
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr1_-_205290756
|
7.401
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr5_-_39270708
|
6.942
|
NM_001243093
|
FYB
|
FYN binding protein
|
|
chr17_-_43502992
|
6.692
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr12_-_53074172
|
6.392
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr2_-_219031646
|
6.199
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr9_+_135894806
|
6.094
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr6_+_167536240
|
5.901
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr1_-_114414245
|
5.657
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr17_+_25958217
|
5.584
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr1_-_200992510
|
5.560
|
|
KIF21B
|
kinesin family member 21B
|
|
chr9_-_37034358
|
5.321
|
NM_016734
|
PAX5
|
paired box 5
|
|
chr17_+_25958245
|
5.228
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr11_-_111249902
|
5.169
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr17_+_34538464
|
5.134
|
NM_207007
NM_001001435
|
CCL4L2
CCL4L1
|
chemokine (C-C motif) ligand 4-like 2
chemokine (C-C motif) ligand 4-like 1
|
|
chrX_-_19688991
|
5.007
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr6_-_74229094
|
4.844
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr17_+_25958151
|
4.680
|
NM_002308
NM_009587
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr17_+_25958192
|
4.629
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr5_+_96212167
|
4.532
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr2_-_152146373
|
4.451
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr11_+_69061599
|
4.328
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chrX_+_37639306
|
4.175
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr1_-_26633110
|
3.934
|
NM_001077262
NM_183008
|
UBXN11
|
UBX domain protein 11
|
|
chr14_-_106994001
|
3.845
|
|
IGHV3-48
IGH@
IGHA1
IGHM
|
immunoglobulin heavy variable 3-48
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr14_-_106692107
|
3.808
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG3
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr17_+_25958257
|
3.700
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr19_-_17958773
|
3.645
|
|
JAK3
|
Janus kinase 3
|
|
chr2_-_100722328
|
3.594
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr16_-_67965754
|
3.391
|
NM_001907
|
CTRL
|
chymotrypsin-like
|
|
chr5_+_96211643
|
3.353
|
NM_001130140
NM_022350
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr16_+_12058963
|
3.301
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr14_-_24809031
|
3.216
|
NM_006871
|
RIPK3
|
receptor-interacting serine-threonine kinase 3
|
|
chr1_-_27953046
|
3.146
|
NM_001042729
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr7_-_36764000
|
3.025
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr12_+_69742140
|
2.969
|
|
LYZ
|
lysozyme
|
|
chr14_-_106714594
|
2.884
|
|
|
|
|
chr9_+_26066672
|
2.860
|
NM_001004352
|
LOC100506422
|
putative deoxyuridine 5'-triphosphate nucleotidohydrolase-like protein FLJ16323
|
|
chr19_-_44174497
|
2.830
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr12_-_1920892
|
2.628
|
|
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4
|
|
chr17_+_56414563
|
2.616
|
|
LOC100506779
|
uncharacterized LOC100506779
|
|
chr9_-_117150217
|
2.511
|
|
AKNA
|
AT-hook transcription factor
|
|
chr16_+_69220940
|
2.509
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr14_-_107218786
|
2.506
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr2_-_37544165
|
2.495
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr14_+_75894508
|
2.470
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr3_+_137728949
|
2.467
|
NM_016369
|
CLDN18
|
claudin 18
|
|
chr3_+_47029453
|
2.419
|
|
NBEAL2
|
neurobeachin-like 2
|
|
chrY_+_21729214
|
2.414
|
|
TXLNG2P
|
taxilin gamma 2, pseudogene
|
|
chr19_+_17638064
|
2.337
|
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr14_-_106725620
|
2.287
|
|
IGHV4-31
IGHV3-23
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-23
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr12_-_7848324
|
2.271
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr19_-_13947101
|
2.233
|
|
LOC284454
|
uncharacterized LOC284454
|
|
chr10_+_70847817
|
2.225
|
NM_002727
|
SRGN
|
serglycin
|
|
chr22_+_22676814
|
2.222
|
|
|
|
|
chr5_-_138725544
|
2.214
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chr4_+_74719012
|
2.202
|
NM_002620
|
PF4V1
|
platelet factor 4 variant 1
|
|
chr7_-_28220342
|
2.129
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr5_+_35856990
|
2.104
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr5_-_39203061
|
2.095
|
|
FYB
|
FYN binding protein
|
|
chr12_+_69742130
|
2.074
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr10_+_70847857
|
2.060
|
|
SRGN
|
serglycin
|
|
chrX_+_16804531
|
2.060
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr20_+_35973083
|
1.999
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr7_-_81399286
|
1.983
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr14_-_106725232
|
1.903
|
|
ZCWPW2
IGHV3-23
IGKV3-20
IGK@
|
zinc finger, CW type with PWWP domain 2
immunoglobulin heavy variable 3-23
immunoglobulin kappa variable 3-20
immunoglobulin kappa locus
|
|
chr11_-_59950498
|
1.858
|
NM_022349
NM_152851
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A
|
|
chr11_-_66112537
|
1.855
|
NM_001024957
NM_015399
|
BRMS1
|
breast cancer metastasis suppressor 1
|
|
chr11_+_60223281
|
1.841
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr1_-_161039586
|
1.840
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr6_-_97731051
|
1.827
|
NM_198468
|
MMS22L
|
MMS22-like, DNA repair protein
|
|
chr1_+_192127591
|
1.820
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr17_+_7461591
|
1.747
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr11_-_44972227
|
1.730
|
NM_001076787
NM_006034
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr8_-_145060563
|
1.698
|
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr19_+_55281262
|
1.679
|
NM_014218
|
KIR2DL1
KIR2DS4
|
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 1
killer cell immunoglobulin-like receptor, two domains, short cytoplasmic tail, 4
|
|
chr1_+_28585959
|
1.677
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr8_-_145060594
|
1.661
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr11_+_122709205
|
1.661
|
NM_019604
|
CRTAM
|
cytotoxic and regulatory T cell molecule
|
|
chr14_-_107078578
|
1.625
|
|
|
|
|
chr14_-_106691698
|
1.603
|
|
|
|
|
chr1_+_174844655
|
1.592
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr11_-_111750144
|
1.588
|
NM_138378
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1
|
|
chr1_-_161039413
|
1.553
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr19_-_54311958
|
1.548
|
NM_033297
|
NLRP12
|
NLR family, pyrin domain containing 12
|
|
chr1_-_12908141
|
1.518
|
NM_001146181
|
HNRNPCL1
LOC649330
|
heterogeneous nuclear ribonucleoprotein C-like 1
heterogeneous nuclear ribonucleoprotein C-like
|
|
chr11_+_68857483
|
1.500
|
|
|
|
|
chr5_+_14664764
|
1.463
|
NM_138348
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr17_-_40306988
|
1.446
|
|
RAB5C
|
RAB5C, member RAS oncogene family
|
|
chr4_-_5711274
|
1.441
|
NM_001166136
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr12_-_120687956
|
1.425
|
NM_025157
|
PXN
|
paxillin
|
|
chr10_-_23003443
|
1.418
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr2_+_171571936
|
1.373
|
|
|
|
|
chr4_+_155484131
|
1.364
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr9_+_75766777
|
1.346
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr2_+_171571800
|
1.342
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr3_-_10332408
|
1.316
|
NM_001134941
NM_016362
|
GHRL
|
ghrelin/obestatin prepropeptide
|
|
chr17_-_40306935
|
1.294
|
|
RAB5C
|
RAB5C, member RAS oncogene family
|
|
chr17_-_79881304
|
1.293
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr11_-_111749810
|
1.239
|
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1
|
|
chr3_+_128532267
|
1.236
|
|
|
|
|
chr20_-_50179338
|
1.220
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr16_-_4323055
|
1.207
|
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr4_-_151936648
|
1.198
|
NM_006726
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr19_+_2289996
|
1.194
|
|
|
|
|
chr7_+_143013218
|
1.183
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr10_+_135051407
|
1.178
|
NM_014468
|
VENTX
|
VENT homeobox
|
|
chr7_-_5570188
|
1.175
|
NM_001101
|
ACTB
|
actin, beta
|
|
chr9_+_75766794
|
1.162
|
|
ANXA1
|
annexin A1
|
|
chr16_+_77233348
|
1.161
|
NM_001129979
|
SYCE1L
|
synaptonemal complex central element protein 1-like
|
|
chr19_-_11688508
|
1.142
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr10_-_125853102
|
1.116
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr15_-_80263460
|
1.110
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr1_+_27101217
|
1.103
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr4_-_151936864
|
1.096
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chrX_+_57313109
|
1.084
|
NM_174912
|
FAAH2
|
fatty acid amide hydrolase 2
|
|
chr7_+_17382900
|
1.083
|
|
AHR
|
aryl hydrocarbon receptor
|
|
chr2_+_219246751
|
1.077
|
NM_000578
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1
|
|
chr17_+_56406294
|
1.070
|
|
LOC100506779
|
uncharacterized LOC100506779
|
|
chr16_-_4323000
|
1.067
|
NM_003223
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr19_-_58864820
|
1.063
|
NM_130786
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr1_-_1141932
|
1.057
|
NM_004195
NM_148901
NM_148902
|
TNFRSF18
|
tumor necrosis factor receptor superfamily, member 18
|
|
chr19_+_7828034
|
1.055
|
NM_001144904
NM_001144905
NM_001144906
NM_001144907
NM_001144908
NM_001144909
NM_001144910
NM_001144911
NM_014257
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr15_-_40600025
|
1.005
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr15_-_31523049
|
0.995
|
NM_001243538
|
LOC283710
|
uncharacterized LOC283710
|
|
chr17_+_57970442
|
0.992
|
NM_003161
|
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1
|
|
chr6_+_109762204
|
0.986
|
|
SMPD2
|
sphingomyelin phosphodiesterase 2, neutral membrane (neutral sphingomyelinase)
|
|
chr17_+_34640030
|
0.984
|
NM_207007
NM_001001435
|
CCL4L2
CCL4L1
|
chemokine (C-C motif) ligand 4-like 2
chemokine (C-C motif) ligand 4-like 1
|
|
chr20_-_52199635
|
0.982
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr4_+_153864134
|
0.976
|
NM_033393
|
FHDC1
|
FH2 domain containing 1
|
|
chr5_-_49737198
|
0.972
|
|
EMB
|
embigin
|
|
chr19_-_11688441
|
0.952
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr5_+_14664803
|
0.947
|
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr10_+_111765725
|
0.927
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr9_+_141106636
|
0.924
|
NM_001145249
|
FAM157B
|
family with sequence similarity 157, member B
|
|
chr21_-_15918635
|
0.923
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr6_-_128222175
|
0.923
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr6_+_132873829
|
0.922
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr20_-_62199106
|
0.917
|
NM_033405
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr1_+_39456887
|
0.895
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr1_+_45266048
|
0.875
|
|
PLK3
|
polo-like kinase 3
|
|
chr1_+_66797775
|
0.873
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr6_-_32634287
|
0.871
|
NM_001243961
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr8_-_143833924
|
0.857
|
NM_205545
|
LYPD2
|
LY6/PLAUR domain containing 2
|
|
chr6_-_6320875
|
0.846
|
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr17_-_40307015
|
0.846
|
NM_001252039
NM_004583
NM_201434
|
RAB5C
|
RAB5C, member RAS oncogene family
|
|
chr19_-_44174271
|
0.845
|
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr8_-_119123804
|
0.839
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr7_+_99816870
|
0.833
|
NM_024070
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing
|
|
chr5_-_49737216
|
0.832
|
NM_198449
|
EMB
|
embigin
|
|
chr12_+_56324945
|
0.826
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr16_-_88717395
|
0.818
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr19_-_44285012
|
0.813
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr5_-_138718941
|
0.813
|
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr6_-_31681845
|
0.811
|
|
|
|
|
chr1_-_40562917
|
0.809
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr8_+_22019167
|
0.789
|
NM_001172357
NM_001172410
NM_003018
|
SFTPC
|
surfactant protein C
|
|
chr17_+_7462064
|
0.770
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr2_+_228678556
|
0.767
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr3_-_71542628
|
0.766
|
|
FOXP1
|
forkhead box P1
|
|
chr1_-_205601999
|
0.753
|
NM_001973
NM_021795
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr16_+_56995761
|
0.738
|
NM_000078
|
CETP
|
cholesteryl ester transfer protein, plasma
|
|
chr17_+_1619816
|
0.726
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr14_-_51411118
|
0.715
|
NM_001163940
NM_002863
|
PYGL
|
phosphorylase, glycogen, liver
|
|
chr9_-_130533509
|
0.709
|
NM_001252334
NM_005489
|
SH2D3C
|
SH2 domain containing 3C
|
|
chrY_+_24636543
|
0.682
|
NM_001002758
NM_004676
|
PRY2
PRY
|
PTPN13-like, Y-linked 2
PTPN13-like, Y-linked
|
|
chr7_+_72396799
|
0.673
|
|
POM121
|
POM121 membrane glycoprotein
|
|
chr1_-_40562909
|
0.672
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr1_+_156123321
|
0.663
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr19_+_11546085
|
0.656
|
NM_001001329
NM_002743
|
PRKCSH
|
protein kinase C substrate 80K-H
|
|
chr10_+_114133838
|
0.654
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr4_+_160188997
|
0.644
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr15_+_91411884
|
0.640
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr1_+_174933904
|
0.639
|
NM_001243764
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr12_-_51663969
|
0.630
|
NM_001033873
NM_001031628
|
SMAGP
|
small cell adhesion glycoprotein
|
|
chr15_-_34502269
|
0.625
|
NM_024713
|
C15orf29
|
chromosome 15 open reading frame 29
|
|
chr10_+_11047253
|
0.612
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chrY_-_24242153
|
0.611
|
NM_001002758
NM_004676
|
PRY2
PRY
|
PTPN13-like, Y-linked 2
PTPN13-like, Y-linked
|
|
chr1_+_45266035
|
0.610
|
NM_004073
|
PLK3
|
polo-like kinase 3
|
|
chr3_-_58200397
|
0.589
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr17_-_61523544
|
0.584
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr11_+_60524339
|
0.583
|
NM_001098835
NM_152717
|
MS4A15
|
membrane-spanning 4-domains, subfamily A, member 15
|
|
chr12_+_96252705
|
0.582
|
NM_003095
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr19_+_55174123
|
0.575
|
NM_001081438
NM_006847
|
LILRB4
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4
|
|
chr11_+_129685732
|
0.574
|
NM_138788
|
TMEM45B
|
transmembrane protein 45B
|
|
chr17_+_7482961
|
0.572
|
|
CD68
|
CD68 molecule
|
|
chr17_+_7482938
|
0.571
|
|
CD68
|
CD68 molecule
|
|
chr20_+_825284
|
0.566
|
NM_031424
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chrX_+_23685658
|
0.561
|
|
PRDX4
|
peroxiredoxin 4
|
|
chr3_+_52812438
|
0.558
|
NM_001166434
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr11_+_59856136
|
0.556
|
NM_000139
|
MS4A2
|
membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide)
|