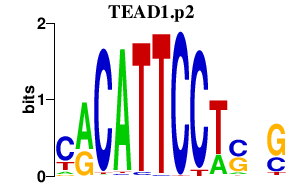
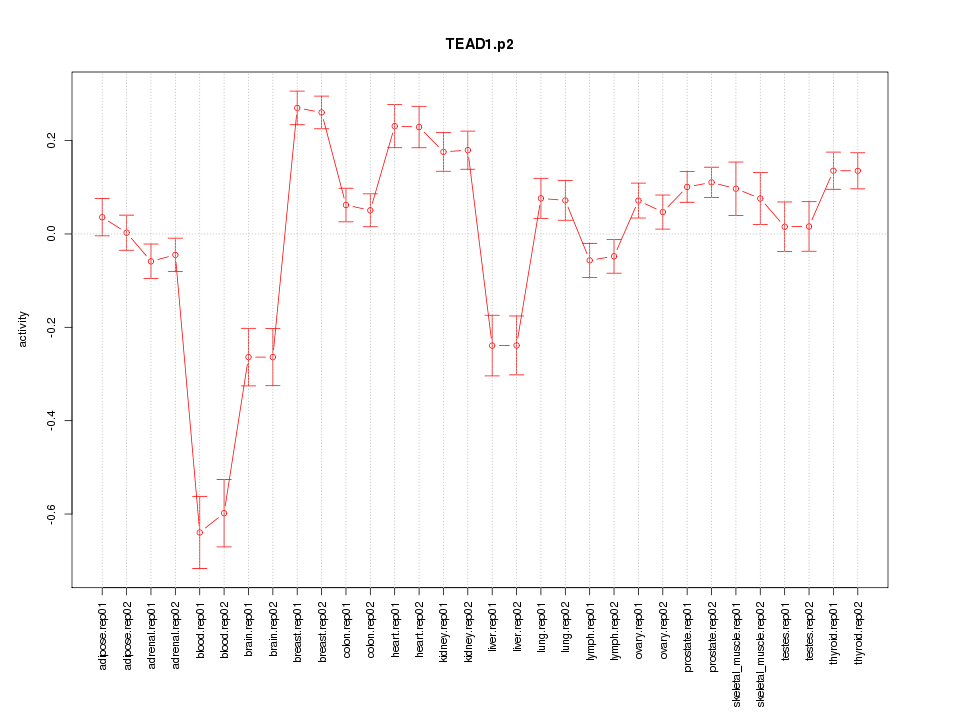
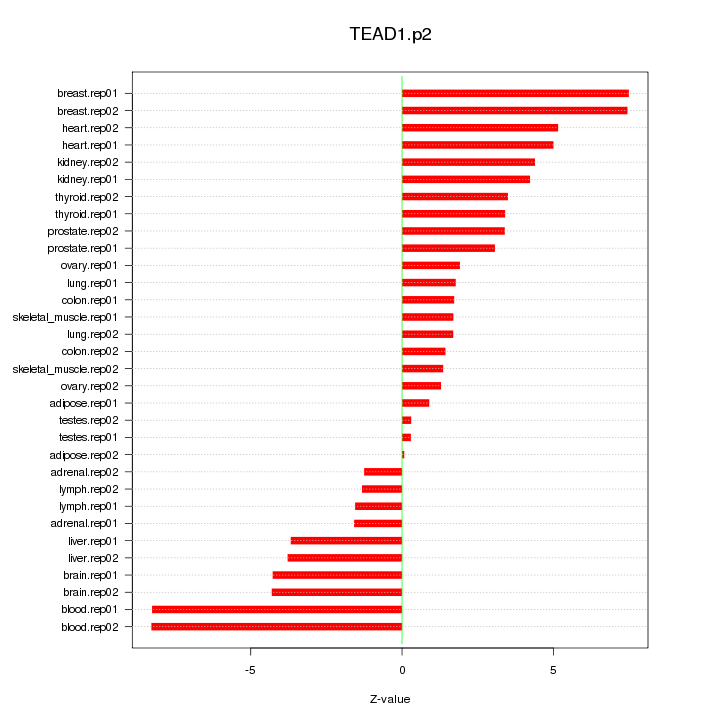
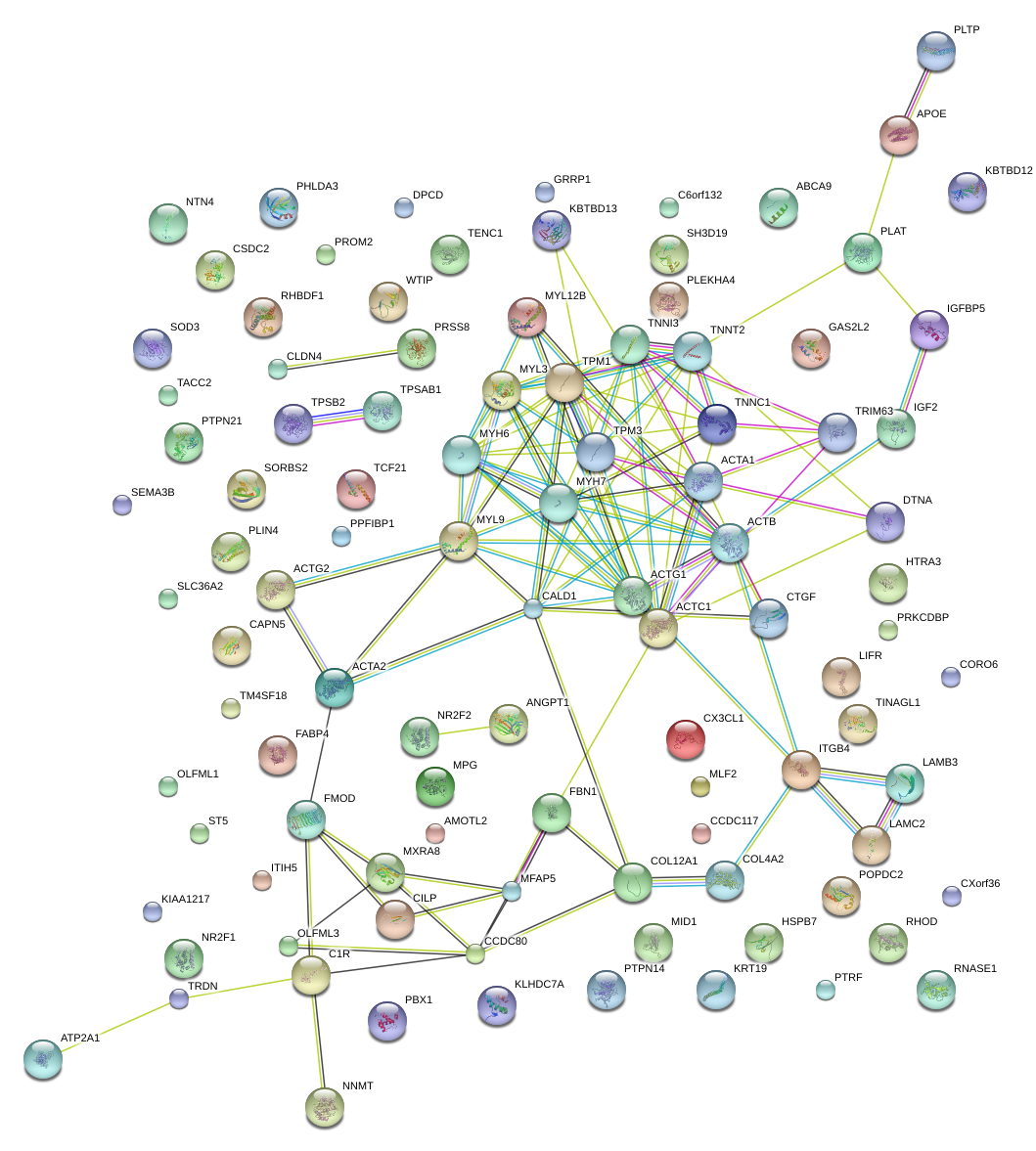
|
chr19_+_34972866
|
34.910
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr6_-_132272311
|
27.080
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr1_-_214724974
|
24.143
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr3_-_119379269
|
19.348
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr1_-_16344438
|
18.233
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr1_-_214724844
|
17.992
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr1_-_203317366
|
16.513
|
|
FMOD
|
fibromodulin
|
|
chr3_-_52488030
|
15.273
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chr2_-_217540722
|
15.264
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr14_-_23904849
|
14.375
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr5_-_138842291
|
13.647
|
NM_001077693
|
ECSCR
|
endothelial cell-specific chemotaxis regulator
|
|
chr10_-_90712510
|
13.566
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr17_-_40575117
|
13.485
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr4_-_186733372
|
12.972
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_+_24797203
|
12.840
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr3_-_112359962
|
12.618
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr15_+_63335086
|
12.242
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr1_-_26393984
|
12.007
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr6_-_75915566
|
11.519
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr3_-_112359443
|
11.425
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr12_-_7245006
|
11.404
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr3_-_112359660
|
11.361
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359863
|
11.290
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr11_+_66824308
|
11.245
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr1_-_1293907
|
11.115
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr15_-_65503786
|
10.848
|
NM_003613
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase
|
|
chr3_-_112359277
|
10.839
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359251
|
10.766
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359565
|
10.744
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_26485510
|
10.486
|
NM_024869
|
FAM110D
|
family with sequence similarity 110, member D
|
|
chr1_+_114522029
|
10.483
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr16_-_122573
|
10.260
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr6_-_42110714
|
10.227
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr11_+_66824285
|
10.016
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr3_-_112359547
|
10.006
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_32043745
|
9.950
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr11_-_6341708
|
9.905
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr12_+_53442732
|
9.870
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr12_-_8815370
|
9.807
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr15_+_63334753
|
9.753
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr10_+_123872440
|
9.688
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr19_-_49371710
|
9.614
|
NM_001161354
NM_020904
|
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4
|
|
chr4_-_186578122
|
9.587
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_183155393
|
9.579
|
|
LAMC2
|
laminin, gamma 2
|
|
chr3_-_149051258
|
9.518
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr6_-_123957941
|
9.510
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr16_-_31146992
|
9.466
|
|
PRSS8
|
protease, serine, 8
|
|
chr11_+_7506599
|
9.457
|
NM_198474
|
OLFML1
|
olfactomedin-like 1
|
|
chr1_-_201346804
|
9.397
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr17_-_39684499
|
9.371
|
|
KRT19
|
keratin 19
|
|
chr22_+_41957011
|
9.305
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr16_+_28889808
|
9.243
|
NM_004320
NM_173201
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr4_-_152149042
|
9.234
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr10_+_24498088
|
9.018
|
|
KIAA1217
|
KIAA1217
|
|
chr14_-_23877480
|
9.001
|
NM_002471
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha
|
|
chr10_-_7682741
|
8.999
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr12_-_7245048
|
8.898
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr5_-_38595506
|
8.846
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr12_-_96184510
|
8.808
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr14_-_89020888
|
8.657
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr16_+_57406398
|
8.476
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr11_+_114166534
|
8.460
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr17_-_27949814
|
8.353
|
|
CORO6
|
coronin 6
|
|
chr15_+_96869156
|
8.192
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chrX_-_45060081
|
8.098
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr12_-_8815266
|
8.060
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr17_-_67057046
|
8.003
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr2_+_95940195
|
7.968
|
NM_001165977
NM_001165978
NM_144707
|
PROM2
|
prominin 2
|
|
chr1_+_183155361
|
7.934
|
|
LAMC2
|
laminin, gamma 2
|
|
chr20_-_44540685
|
7.846
|
|
PLTP
|
phospholipid transfer protein
|
|
chr3_+_127641901
|
7.840
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr8_-_82395401
|
7.586
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr19_+_45409003
|
7.544
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr17_-_34079896
|
7.540
|
NM_139285
|
GAS2L2
|
growth arrest-specific 2 like 2
|
|
chr1_-_209825673
|
7.410
|
NM_001127641
|
LAMB3
|
laminin, beta 3
|
|
chr3_-_46904879
|
7.369
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr3_+_50306578
|
7.265
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr1_-_209825797
|
7.245
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr3_-_134093235
|
7.236
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr11_-_8832881
|
7.213
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr1_-_201438150
|
7.060
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr17_+_73717556
|
7.050
|
|
ITGB4
|
integrin, beta 4
|
|
chr6_+_134210264
|
6.994
|
|
TCF21
|
transcription factor 21
|
|
chrX_-_10588458
|
6.991
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr10_+_24497719
|
6.977
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr7_+_73245192
|
6.916
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr6_+_134210247
|
6.859
|
NM_198392
NM_003206
|
TCF21
|
transcription factor 21
|
|
chr1_+_164528586
|
6.852
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr8_-_108510094
|
6.810
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr1_-_201346783
|
6.802
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr1_+_183155173
|
6.788
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr4_+_8271458
|
6.720
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr19_-_4517715
|
6.715
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr11_-_2170792
|
6.707
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr10_+_123748688
|
6.566
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr14_-_21270537
|
6.560
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr15_-_48937795
|
6.534
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr7_-_142985270
|
6.454
|
|
|
|
|
chr12_+_27677044
|
6.439
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr18_+_32173281
|
6.383
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr5_-_150727088
|
6.368
|
NM_181776
|
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2
|
|
chr15_+_65369153
|
6.359
|
NM_001101362
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13
|
|
chr12_-_8815479
|
6.347
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr14_-_21271002
|
6.323
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr16_-_31147062
|
6.157
|
NM_002773
|
PRSS8
|
protease, serine, 8
|
|
chr1_+_18807423
|
6.119
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr16_-_1280162
|
6.112
|
NM_024164
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr8_-_42065037
|
6.056
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr20_+_35169876
|
6.002
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr11_-_8832189
|
5.995
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr7_+_134464372
|
5.993
|
|
CALD1
|
caldesmon 1
|
|
chr13_+_111138079
|
5.977
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr8_+_70378858
|
5.927
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr1_-_115238091
|
5.906
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr14_-_23445885
|
5.885
|
|
AJUBA
|
ajuba LIM protein
|
|
chr3_+_50306305
|
5.862
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr11_-_47470687
|
5.857
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr16_-_31439650
|
5.815
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr17_+_45286713
|
5.770
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr18_-_21242592
|
5.769
|
NM_173505
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr20_+_35169897
|
5.760
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr1_+_78354199
|
5.631
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr7_+_134464161
|
5.621
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr2_+_228029280
|
5.615
|
NM_000091
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen)
|
|
chr12_-_57634474
|
5.512
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr2_-_190044330
|
5.498
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr8_+_1772095
|
5.473
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr1_+_86046434
|
5.407
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr8_-_12973752
|
5.386
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr19_+_50936159
|
5.239
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr10_+_50507186
|
5.155
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr17_+_72429835
|
5.137
|
NM_018653
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr6_-_30899926
|
5.081
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr9_+_137533713
|
5.020
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr10_-_21186530
|
5.010
|
NM_006393
|
NEBL
|
nebulette
|
|
chr14_-_23446431
|
5.004
|
NM_198086
|
AJUBA
|
ajuba LIM protein
|
|
chr12_+_3068442
|
4.995
|
NM_003213
NM_201441
NM_201443
|
TEAD4
|
TEA domain family member 4
|
|
chr2_+_102759184
|
4.989
|
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr12_-_56101465
|
4.988
|
|
ITGA7
|
integrin, alpha 7
|
|
chr6_-_75915753
|
4.980
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr7_+_134464417
|
4.960
|
|
CALD1
|
caldesmon 1
|
|
chr17_-_39684585
|
4.952
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr16_+_1290677
|
4.819
|
NM_003294
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr4_+_184020343
|
4.736
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr1_-_24438625
|
4.723
|
NM_152372
|
MYOM3
|
myomesin family, member 3
|
|
chr11_-_76380894
|
4.720
|
NM_005512
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr16_+_57406382
|
4.712
|
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr6_-_168479796
|
4.665
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr17_+_4901199
|
4.653
|
NM_006612
|
KIF1C
|
kinesin family member 1C
|
|
chr3_+_9944511
|
4.641
|
NM_001193380
NM_153480
|
IL17RE
|
interleukin 17 receptor E
|
|
chr16_+_30907927
|
4.598
|
NM_001142544
NM_001330
|
CTF1
|
cardiotrophin 1
|
|
chr15_-_99548790
|
4.596
|
NM_001102612
|
PGPEP1L
|
pyroglutamyl-peptidase I-like
|
|
chr19_+_50691437
|
4.559
|
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr12_-_80084742
|
4.553
|
NM_002583
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr17_-_40828078
|
4.491
|
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr3_+_28431986
|
4.461
|
NM_001040432
|
ZCWPW2
|
zinc finger, CW type with PWWP domain 2
|
|
chrX_-_31526414
|
4.454
|
NM_004014
|
DMD
|
dystrophin
|
|
chr3_+_190105660
|
4.414
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr8_-_42065193
|
4.380
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr3_+_9944295
|
4.375
|
NM_153483
|
IL17RE
|
interleukin 17 receptor E
|
|
chr7_+_116164838
|
4.330
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr9_-_95166827
|
4.314
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr17_+_73717515
|
4.268
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr10_-_88730928
|
4.249
|
|
LOC100133190
|
uncharacterized LOC100133190
|
|
chr17_+_73717596
|
4.208
|
|
ITGB4
|
integrin, beta 4
|
|
chr19_+_45349584
|
4.136
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr2_-_238499713
|
4.133
|
NM_022449
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr6_-_53530505
|
4.130
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr17_+_9729380
|
4.104
|
NM_004246
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr2_-_216246961
|
4.101
|
|
FN1
|
fibronectin 1
|
|
chr16_+_4838397
|
4.069
|
NM_001253790
NM_001253791
|
LOC440335
|
uncharacterized LOC440335
|
|
chrX_+_114795409
|
4.015
|
|
PLS3
|
plastin 3
|
|
chr19_-_51456159
|
4.013
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr3_+_136537860
|
3.986
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr11_-_46939908
|
3.986
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr12_-_56101667
|
3.966
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr15_-_69707358
|
3.920
|
|
LOC145694
|
uncharacterized LOC145694
|
|
chr19_-_36822597
|
3.887
|
|
LOC100506930
|
uncharacterized LOC100506930
|
|
chr6_+_30850693
|
3.875
|
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr11_-_119217365
|
3.861
|
NM_015645
NM_031433
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr4_+_41540320
|
3.781
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_41510627
|
3.697
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr3_-_49941300
|
3.691
|
NM_001244937
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr16_-_46782104
|
3.690
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr17_+_37856174
|
3.677
|
|
|
|
|
chr17_+_48912604
|
3.651
|
NM_175575
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2
|
|
chr12_+_54410893
|
3.584
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr17_+_7758380
|
3.558
|
NM_203411
|
TMEM88
|
transmembrane protein 88
|
|
chr9_+_118916069
|
3.552
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr12_+_26348488
|
3.521
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr3_-_52869234
|
3.501
|
NM_205853
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr19_-_55580896
|
3.494
|
NM_138412
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr22_+_32439103
|
3.487
|
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1
|
|
chr14_+_95047730
|
3.483
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr11_+_1855673
|
3.477
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chr16_-_68002455
|
3.474
|
NM_001145961
NM_001145962
NM_005072
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr1_+_20396652
|
3.467
|
NM_000929
|
PLA2G5
|
phospholipase A2, group V
|
|
chr2_-_238499317
|
3.451
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr1_-_161279724
|
3.448
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr17_-_48278987
|
3.439
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|