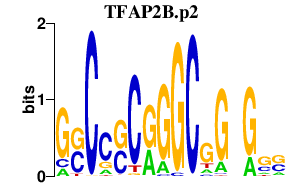
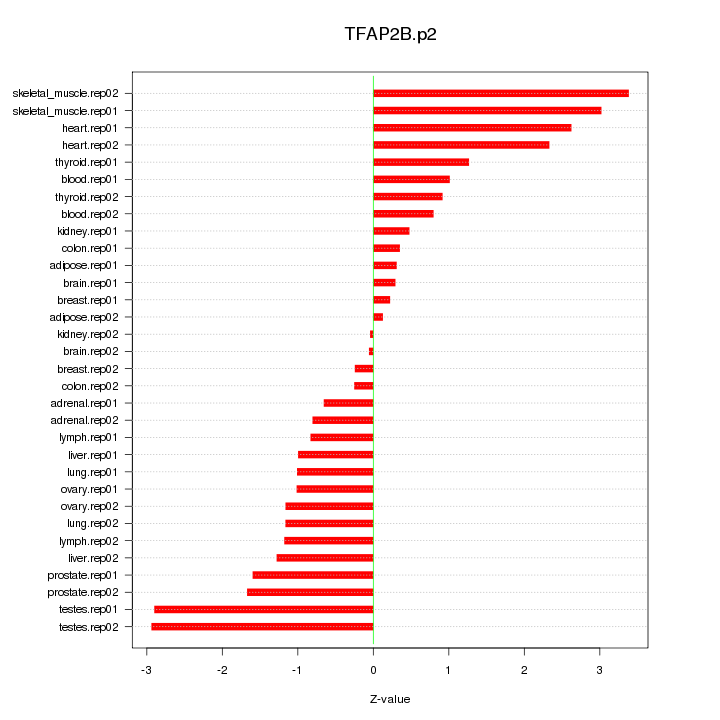
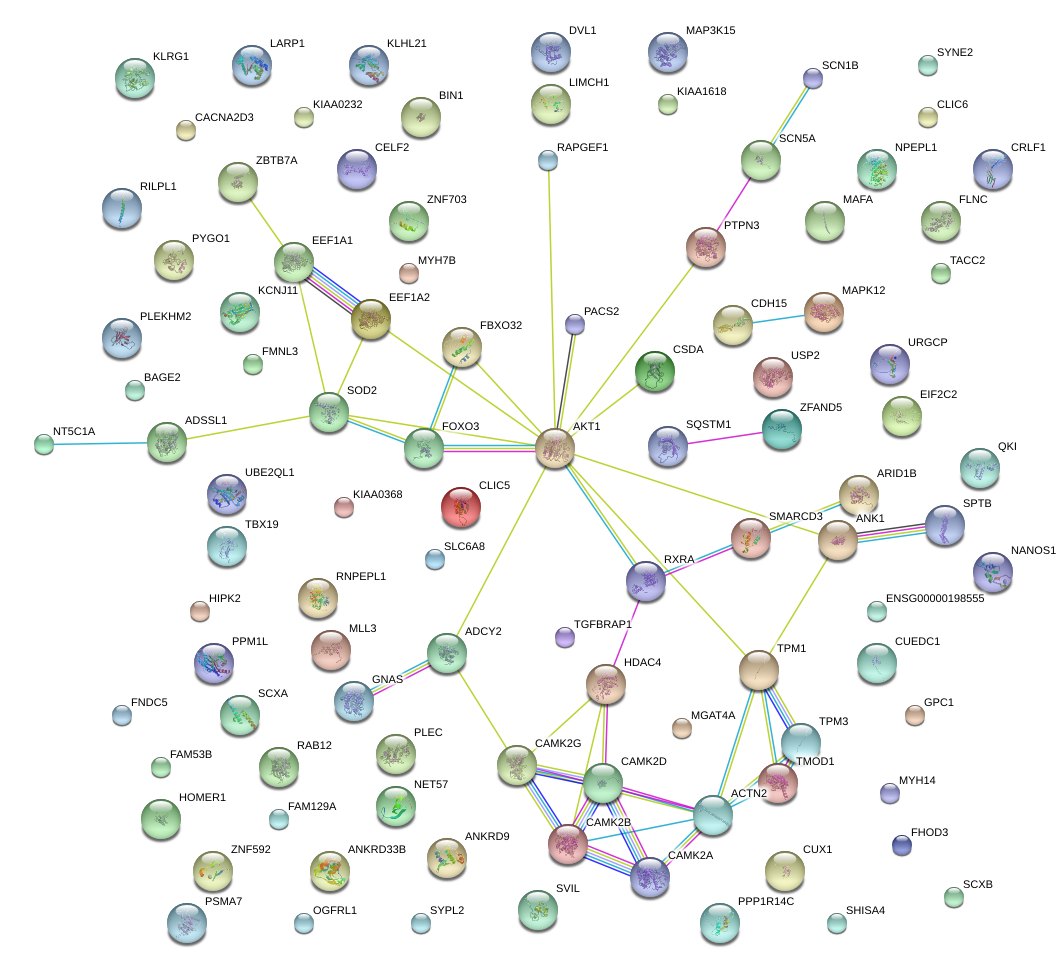
|
chr6_-_160114318
|
3.255
|
NM_000636
NM_001024465
NM_001024466
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr6_-_160114186
|
3.124
|
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr20_-_62130390
|
2.866
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr8_-_124553448
|
2.600
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr8_-_141645603
|
2.410
|
NM_001164623
NM_012154
|
EIF2C2
|
eukaryotic translation initiation factor 2C, 2
|
|
chr5_+_7396030
|
2.264
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr17_+_78234626
|
2.260
|
NM_020914
NM_020954
|
RNF213
|
ring finger protein 213
|
|
chr2_+_241508103
|
2.219
|
|
RNPEPL1
|
arginyl aminopeptidase (aminopeptidase B)-like 1
|
|
chr3_-_38691118
|
2.177
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr1_+_236849753
|
2.135
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr2_+_241507928
|
2.127
|
NM_018226
|
RNPEPL1
|
arginyl aminopeptidase (aminopeptidase B)-like 1
|
|
chr20_+_57466509
|
2.114
|
|
GNAS
|
GNAS complex locus
|
|
chr2_-_240322620
|
2.108
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr2_-_240322692
|
2.096
|
|
HDAC4
|
histone deacetylase 4
|
|
chr22_-_50699938
|
2.074
|
NM_002969
|
MAPK12
|
mitogen-activated protein kinase 12
|
|
chr20_+_57466248
|
2.072
|
NM_000516
NM_001077488
NM_001077489
NM_080426
|
GNAS
|
GNAS complex locus
|
|
chr6_+_157099262
|
2.047
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr1_-_33336289
|
2.009
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr5_-_78809658
|
2.003
|
NM_004272
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr7_-_150946031
|
1.968
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr7_-_44365019
|
1.952
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr2_-_127864505
|
1.951
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr14_+_105190533
|
1.928
|
NM_152328
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr5_+_154135029
|
1.926
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr6_+_108882104
|
1.906
|
|
FOXO3
|
forkhead box O3
|
|
chr7_+_128470462
|
1.883
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr9_+_137218324
|
1.877
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chrX_+_152953751
|
1.835
|
NM_001142805
NM_005629
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr2_+_241375207
|
1.820
|
|
GPC1
|
glypican 1
|
|
chr6_+_157099203
|
1.816
|
|
|
|
|
chr6_+_157099041
|
1.804
|
NM_017519
NM_020732
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr15_+_63335086
|
1.778
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr4_+_6784453
|
1.753
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr19_+_50706868
|
1.718
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr10_-_98592186
|
1.715
|
|
|
|
|
chr6_+_163835783
|
1.704
|
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr12_-_10875848
|
1.690
|
NM_001145426
NM_003651
|
CSDA
|
cold shock domain protein A
|
|
chr14_-_105261850
|
1.653
|
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr8_-_144512601
|
1.637
|
NM_201589
|
MAFA
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian)
|
|
chr4_+_6784462
|
1.625
|
|
KIAA0232
|
KIAA0232
|
|
chr14_-_102976096
|
1.614
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr8_+_145321462
|
1.614
|
NM_001008271
NM_001080514
|
SCXA
SCXB
|
scleraxis homolog A (mouse)
scleraxis homolog B (mouse)
|
|
chr20_+_57267818
|
1.602
|
NM_024663
|
NPEPL1
|
aminopeptidase-like 1
|
|
chr19_-_4065467
|
1.595
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr17_-_56032494
|
1.586
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr1_-_184943532
|
1.583
|
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr14_-_102975996
|
1.575
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr5_+_10564431
|
1.572
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr10_+_123922940
|
1.570
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr8_+_145490548
|
1.564
|
NM_001008271
NM_001080514
|
SCXA
SCXB
|
scleraxis homolog A (mouse)
scleraxis homolog B (mouse)
|
|
chr11_-_17410863
|
1.557
|
NM_001166290
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr22_-_50699687
|
1.540
|
|
MAPK12
|
mitogen-activated protein kinase 12
|
|
chr6_+_108882068
|
1.539
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr1_+_16010838
|
1.529
|
|
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr18_+_33877630
|
1.508
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr9_+_137218313
|
1.483
|
NM_002957
|
RXRA
|
retinoid X receptor, alpha
|
|
chr7_+_101459092
|
1.465
|
NM_001202543
NM_001202544
NM_001202545
NM_001913
NM_181500
NM_001202546
|
CUX1
|
cut-like homeobox 1
|
|
chr20_+_57466721
|
1.460
|
|
GNAS
|
GNAS complex locus
|
|
chr6_+_71998514
|
1.459
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr18_+_8609407
|
1.448
|
NM_001025300
|
RAB12
|
RAB12, member RAS oncogene family
|
|
chr5_+_154134853
|
1.447
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr14_+_105781180
|
1.442
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr5_+_179248089
|
1.438
|
|
SQSTM1
|
sequestosome 1
|
|
chr8_-_145050896
|
1.431
|
NM_000445
|
PLEC
|
plectin
|
|
chr6_+_71998476
|
1.428
|
NM_024576
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr6_-_45983279
|
1.426
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr8_+_37553269
|
1.426
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr9_-_114246225
|
1.417
|
|
KIAA0368
|
KIAA0368
|
|
chr11_-_119234628
|
1.413
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_+_110009220
|
1.389
|
|
SYPL2
|
synaptophysin-like 2
|
|
chr3_+_54156573
|
1.386
|
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr9_-_74979625
|
1.386
|
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr10_-_126432618
|
1.376
|
|
FAM53B
|
family with sequence similarity 53, member B
|
|
chr9_+_100263818
|
1.359
|
NM_003275
|
TMOD1
|
tropomodulin 1
|
|
chr15_+_85291850
|
1.350
|
|
ZNF592
|
zinc finger protein 592
|
|
chr10_+_11060069
|
1.349
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr12_-_124018066
|
1.322
|
NM_178314
|
RILPL1
|
Rab interacting lysosomal protein-like 1
|
|
chr5_+_6448735
|
1.306
|
NM_001145161
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr14_-_65346554
|
1.297
|
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr8_-_41754244
|
1.296
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr7_-_152132924
|
1.295
|
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr14_+_105780903
|
1.295
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr1_-_1284297
|
1.291
|
NM_004421
|
DVL1
|
dishevelled, dsh homolog 1 (Drosophila)
|
|
chr9_-_114245815
|
1.287
|
|
KIAA0368
|
KIAA0368
|
|
chr6_+_71998508
|
1.277
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr3_+_160473286
|
1.269
|
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L
|
|
chr1_+_110009099
|
1.258
|
NM_001040709
|
SYPL2
|
synaptophysin-like 2
|
|
chr19_+_35521591
|
1.233
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr14_+_64319607
|
1.224
|
NM_015180
NM_182914
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr18_+_8609670
|
1.221
|
|
|
|
|
chr9_-_112260529
|
1.221
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr15_-_55880508
|
1.219
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr10_+_11060105
|
1.216
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr7_-_139477437
|
1.213
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr4_+_41362843
|
1.210
|
|
|
|
|
chr9_-_134615216
|
1.207
|
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr1_+_16010816
|
1.199
|
NM_015164
|
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr1_-_40137709
|
1.193
|
NM_032526
|
NT5C1A
|
5'-nucleotidase, cytosolic IA
|
|
chr5_+_179234002
|
1.189
|
NM_001142299
|
SQSTM1
|
sequestosome 1
|
|
chr5_+_179247911
|
1.186
|
|
SQSTM1
|
sequestosome 1
|
|
chr6_+_150464156
|
1.172
|
NM_030949
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C
|
|
chrX_-_19533378
|
1.169
|
NM_001001671
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15
|
|
chr7_+_152161119
|
1.159
|
|
LOC100128822
|
uncharacterized LOC100128822
|
|
chr20_-_60718358
|
1.156
|
|
PSMA7
|
proteasome (prosome, macropain) subunit, alpha type, 7
|
|
chr10_-_126432784
|
1.155
|
|
FAM53B
|
family with sequence similarity 53, member B
|
|
chr10_+_120789227
|
1.153
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr2_-_99347430
|
1.149
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr4_+_41362565
|
1.147
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr10_+_123923104
|
1.147
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr16_+_89238160
|
1.138
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr1_-_6662681
|
1.129
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr2_-_105946104
|
1.127
|
NM_001142621
NM_004257
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1
|
|
chr1_+_201857997
|
1.124
|
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr19_-_18717467
|
1.117
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr10_-_30024594
|
1.116
|
NM_003174
|
SVIL
|
supervillin
|
|
chr14_-_99947113
|
1.105
|
|
SETD3
|
SET domain containing 3
|
|
chr21_+_45285026
|
1.101
|
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr7_+_1570321
|
1.101
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr17_-_56065596
|
1.101
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr16_+_11762235
|
1.096
|
NM_003498
|
SNN
|
stannin
|
|
chr16_-_4166185
|
1.094
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr11_+_33279167
|
1.092
|
NM_001048200
NM_005734
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr12_-_93322911
|
1.092
|
|
EEA1
|
early endosome antigen 1
|
|
chr10_-_126432893
|
1.090
|
NM_014661
|
FAM53B
|
family with sequence similarity 53, member B
|
|
chr14_-_31676680
|
1.087
|
NM_015382
|
HECTD1
|
HECT domain containing 1
|
|
chr19_-_14228362
|
1.081
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr5_+_79330990
|
1.078
|
NM_003248
|
THBS4
|
thrombospondin 4
|
|
chr6_-_105627857
|
1.078
|
NM_022361
|
POPDC3
|
popeye domain containing 3
|
|
chr17_-_56065474
|
1.076
|
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr21_+_45285101
|
1.072
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr1_-_13840157
|
1.066
|
NM_001010847
|
LRRC38
|
leucine rich repeat containing 38
|
|
chr14_-_99947080
|
1.061
|
|
SETD3
|
SET domain containing 3
|
|
chr19_-_18717631
|
1.061
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr1_+_214454276
|
1.059
|
NM_020197
|
SMYD2
|
SET and MYND domain containing 2
|
|
chr9_+_134269509
|
1.059
|
|
PRRC2B
|
proline-rich coiled-coil 2B
|
|
chr8_+_61591358
|
1.054
|
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr18_+_67956242
|
1.053
|
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr18_+_11981457
|
1.052
|
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr3_+_49726961
|
1.049
|
|
RNF123
|
ring finger protein 123
|
|
chr8_-_75233461
|
1.044
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr4_+_6784487
|
1.031
|
|
KIAA0232
|
KIAA0232
|
|
chr20_-_32031425
|
1.027
|
NM_003098
|
SNTA1
|
syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component)
|
|
chr8_+_146277830
|
1.026
|
|
C8orf33
|
chromosome 8 open reading frame 33
|
|
chr12_-_93323006
|
1.022
|
|
EEA1
|
early endosome antigen 1
|
|
chr10_+_134210691
|
1.020
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr1_-_40042415
|
1.018
|
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr20_+_48807460
|
1.014
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr1_-_1284755
|
1.014
|
|
DVL1
|
dishevelled, dsh homolog 1 (Drosophila)
|
|
chr13_+_113344354
|
1.013
|
NM_015205
NM_032189
|
ATP11A
|
ATPase, class VI, type 11A
|
|
chr1_-_184943673
|
1.012
|
NM_052966
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr7_+_4722091
|
1.010
|
|
FOXK1
|
forkhead box K1
|
|
chr4_-_8160435
|
1.010
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr10_-_133795311
|
1.007
|
NM_004052
|
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3
|
|
chr1_+_10270680
|
1.002
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr10_-_133795447
|
0.998
|
|
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3
|
|
chr3_+_20081853
|
0.998
|
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr20_-_3766905
|
0.997
|
NM_001810
|
CENPB
|
centromere protein B, 80kDa
|
|
chr6_-_17987693
|
0.996
|
|
KIF13A
|
kinesin family member 13A
|
|
chr8_-_71315487
|
0.992
|
|
NCOA2
|
nuclear receptor coactivator 2
|
|
chr11_-_119234859
|
0.991
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr2_+_173940589
|
0.991
|
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr9_+_96338671
|
0.981
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr15_+_85291521
|
0.981
|
|
ZNF592
|
zinc finger protein 592
|
|
chr1_-_10754464
|
0.978
|
|
CASZ1
|
castor zinc finger 1
|
|
chr12_-_58132028
|
0.972
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr3_-_48130633
|
0.969
|
NM_001134364
NM_002375
NM_030885
|
MAP4
|
microtubule-associated protein 4
|
|
chr5_+_179247903
|
0.966
|
|
SQSTM1
|
sequestosome 1
|
|
chr17_+_4487833
|
0.964
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr6_-_35656610
|
0.960
|
|
FKBP5
|
FK506 binding protein 5
|
|
chr3_+_135684463
|
0.960
|
NM_001190447
NM_002718
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chrX_+_109246338
|
0.960
|
NM_032227
|
TMEM164
|
transmembrane protein 164
|
|
chrX_+_109246350
|
0.957
|
|
TMEM164
|
transmembrane protein 164
|
|
chr2_+_29338207
|
0.955
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr17_-_27916395
|
0.952
|
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr17_+_45608738
|
0.949
|
|
NPEPPS
|
aminopeptidase puromycin sensitive
|
|
chr17_+_80009775
|
0.948
|
|
GPS1
|
G protein pathway suppressor 1
|
|
chrX_-_15872882
|
0.947
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr12_-_124018428
|
0.945
|
|
RILPL1
|
Rab interacting lysosomal protein-like 1
|
|
chr7_-_150945731
|
0.945
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr17_+_80009739
|
0.945
|
NM_004127
NM_212492
|
GPS1
|
G protein pathway suppressor 1
|
|
chr5_+_179247841
|
0.940
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr16_-_3930120
|
0.939
|
NM_001079846
NM_004380
|
CREBBP
|
CREB binding protein
|
|
chr2_-_242212502
|
0.938
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr7_+_140774031
|
0.936
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr17_-_4269610
|
0.932
|
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1
|
|
chr19_-_15443307
|
0.924
|
|
BRD4
|
bromodomain containing 4
|
|
chr12_-_111180730
|
0.922
|
NM_001244974
NM_002710
|
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme
|
|
chr10_+_105726846
|
0.922
|
|
SLK
|
STE20-like kinase
|
|
chr14_-_91282642
|
0.922
|
|
|
|
|
chr13_+_113863926
|
0.921
|
NM_001008895
|
CUL4A
|
cullin 4A
|
|
chr19_+_3359138
|
0.920
|
NM_001245005
NM_205843
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr3_+_49727014
|
0.919
|
|
RNF123
|
ring finger protein 123
|
|
chr7_+_55955129
|
0.918
|
|
ZNF713
|
zinc finger protein 713
|
|
chr14_-_91282760
|
0.916
|
NM_001010854
|
TTC7B
|
tetratricopeptide repeat domain 7B
|
|
chr1_+_64058891
|
0.915
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr17_-_27507195
|
0.914
|
NM_078471
NM_203318
|
MYO18A
|
myosin XVIIIA
|
|
chr20_-_60718407
|
0.913
|
|
PSMA7
|
proteasome (prosome, macropain) subunit, alpha type, 7
|
|
chr3_-_14989399
|
0.912
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr18_+_11981421
|
0.904
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr6_+_108881010
|
0.903
|
NM_201559
|
FOXO3
|
forkhead box O3
|