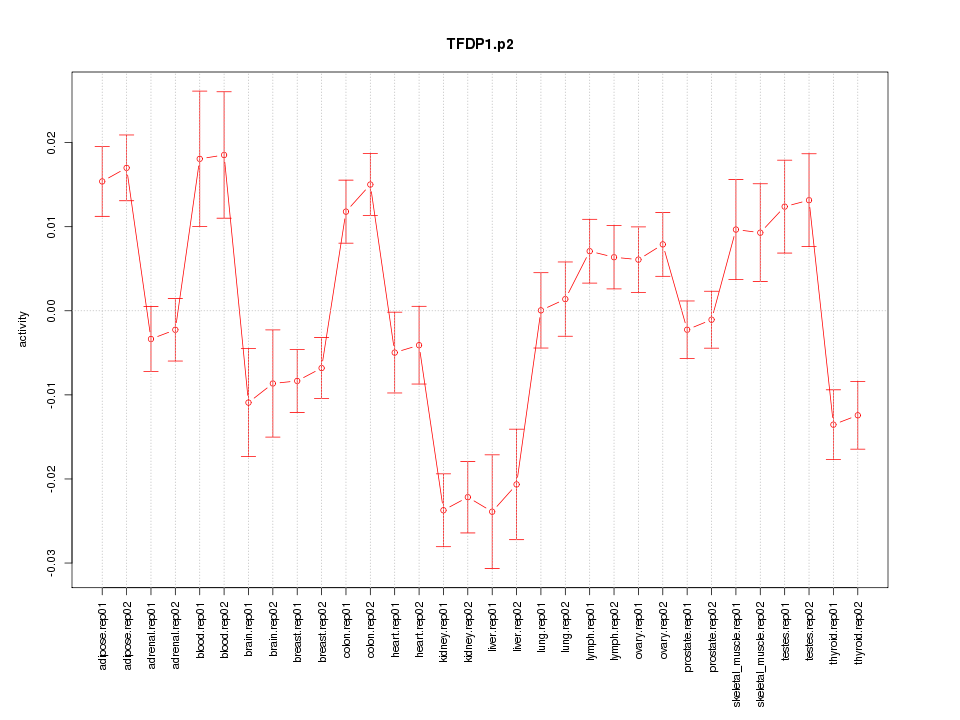
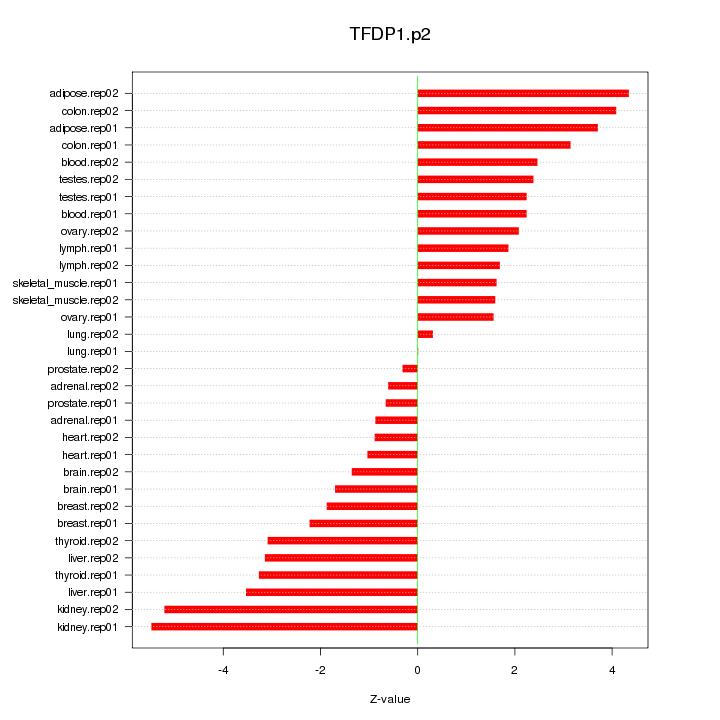
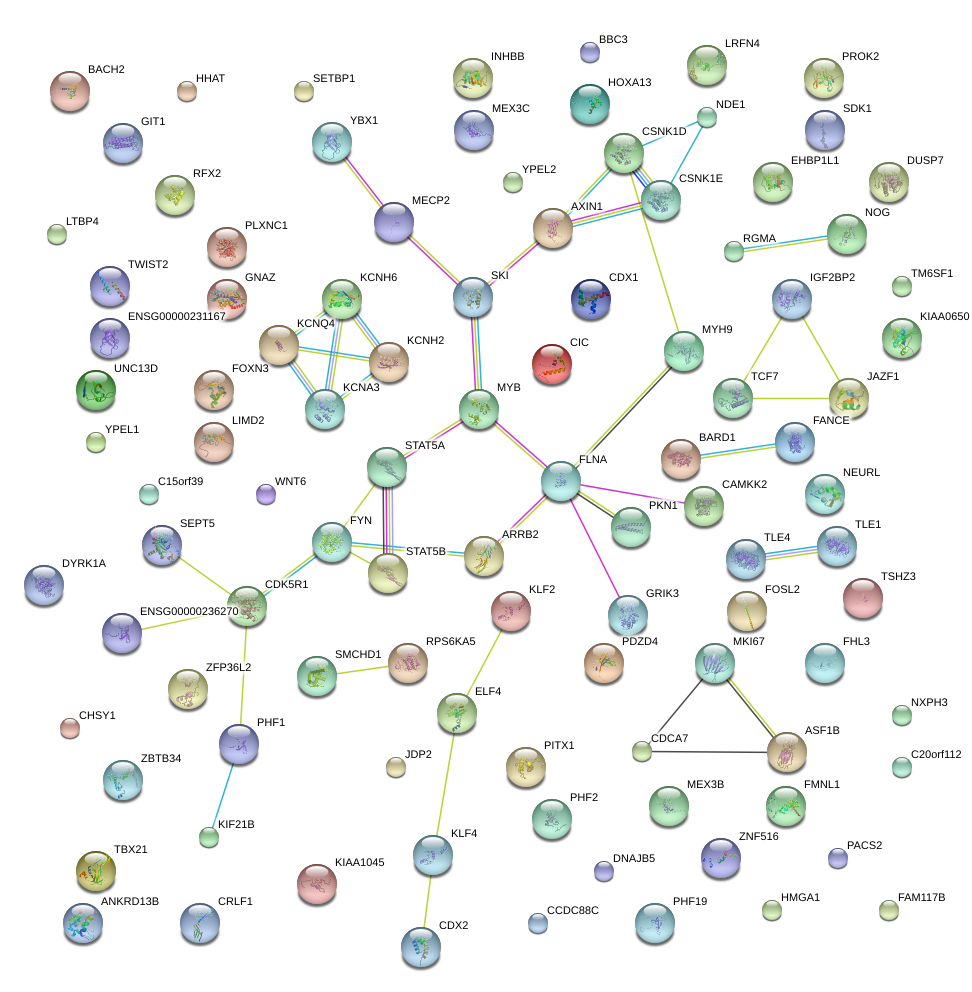
|
chr19_-_47734215
|
3.344
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr3_-_52090090
|
3.333
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr17_-_16395479
|
3.328
|
NM_001113567
NM_207387
|
FAM211A
|
family with sequence similarity 211, member A
|
|
chr6_+_135502521
|
3.291
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr3_-_71834187
|
3.235
|
NM_001126128
NM_021935
|
PROK2
|
prokineticin 2
|
|
chr6_+_34204928
|
3.235
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr5_+_133450353
|
3.218
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr6_+_135502445
|
3.202
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr17_-_16395084
|
3.196
|
|
FAM211A
|
family with sequence similarity 211, member A
|
|
chr6_+_34205029
|
2.995
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr7_-_28220146
|
2.963
|
|
JAZF1
|
JAZF zinc finger 1
|
|
chr14_-_91884164
|
2.928
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr19_-_14247267
|
2.919
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr7_-_27239724
|
2.882
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr9_-_110251754
|
2.838
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr15_-_82338459
|
2.806
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr9_+_82187599
|
2.770
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr2_+_121103670
|
2.747
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr19_-_31840118
|
2.710
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chrX_-_129244652
|
2.706
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr19_-_14247400
|
2.606
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr18_-_74207145
|
2.591
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr19_+_16435734
|
2.530
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr2_-_215674292
|
2.470
|
NM_000465
|
BARD1
|
BRCA1 associated RING domain 1
|
|
chr22_-_36783851
|
2.456
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr14_-_89883306
|
2.403
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr17_+_47653176
|
2.371
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr17_+_47653474
|
2.370
|
|
NXPH3
|
neurexophilin 3
|
|
chr2_+_100721380
|
2.327
|
|
|
|
|
chrX_-_129244471
|
2.316
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr17_+_30813928
|
2.290
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr7_-_150675236
|
2.287
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr9_+_34958171
|
2.274
|
NM_015297
|
KIAA1045
|
KIAA1045
|
|
chr15_+_75494235
|
2.267
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr15_+_75494196
|
2.245
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr19_+_14544100
|
2.225
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr22_-_36784001
|
2.217
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr7_+_3340919
|
2.206
|
NM_152744
|
SDK1
|
sidekick cell adhesion molecule 1
|
|
chr17_-_27916395
|
2.206
|
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr6_+_34204690
|
2.197
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_+_219724545
|
2.190
|
NM_006522
|
WNT6
|
wingless-type MMTV integration site family, member 6
|
|
chr11_+_65343498
|
2.158
|
NM_001099409
|
EHBP1L1
|
EH domain binding protein 1-like 1
|
|
chr22_-_36783930
|
2.143
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr10_+_105253734
|
2.138
|
NM_004210
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr15_+_75494233
|
2.133
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr3_-_185542744
|
2.083
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr17_-_27916620
|
2.083
|
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr19_+_14544216
|
2.065
|
|
PKN1
|
protein kinase N1
|
|
chr22_+_23412364
|
2.030
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr2_+_28616360
|
2.025
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr12_+_94542239
|
1.988
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr5_-_134369571
|
1.988
|
|
PITX1
|
paired-like homeodomain 1
|
|
chr17_+_43299277
|
1.978
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr1_+_2160133
|
1.963
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr19_+_16435632
|
1.960
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr6_-_91006460
|
1.953
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_+_203500211
|
1.938
|
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr19_+_42788605
|
1.937
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr17_+_45810609
|
1.896
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr15_-_101791674
|
1.877
|
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr15_-_82338348
|
1.876
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr15_+_83776323
|
1.850
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chrX_-_153602928
|
1.839
|
|
FLNA
|
filamin A, alpha
|
|
chrX_-_153363143
|
1.835
|
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome)
|
|
chr9_-_110251308
|
1.831
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr17_+_54671059
|
1.804
|
NM_005450
|
NOG
|
noggin
|
|
chr17_+_27920479
|
1.790
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr6_+_34204576
|
1.789
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr22_-_36783977
|
1.787
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr22_-_22089975
|
1.782
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr2_+_239756672
|
1.778
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr6_-_112194648
|
1.764
|
NM_002037
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr16_-_402617
|
1.762
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr9_-_123639566
|
1.761
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr22_+_23412727
|
1.761
|
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr9_+_96338671
|
1.746
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr9_+_129622935
|
1.733
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr1_-_38471170
|
1.729
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr19_-_18717577
|
1.726
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr18_+_2655885
|
1.713
|
NM_015295
|
SMCHD1
|
structural maintenance of chromosomes flexible hinge domain containing 1
|
|
chr17_-_61777467
|
1.709
|
|
LIMD2
|
LIM domain containing 2
|
|
chr12_-_121734474
|
1.699
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr14_-_91526912
|
1.695
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr20_-_31172874
|
1.693
|
|
LOC284804
|
uncharacterized LOC284804
|
|
chr22_+_19701968
|
1.679
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr18_-_48723689
|
1.665
|
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr1_+_43148187
|
1.664
|
|
YBX1
|
Y box binding protein 1
|
|
chr17_-_40428391
|
1.654
|
NM_012448
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr1_-_200992510
|
1.642
|
|
KIF21B
|
kinesin family member 21B
|
|
chr19_-_6110491
|
1.632
|
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr2_+_203499819
|
1.630
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chrX_-_129244422
|
1.627
|
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr19_-_18717631
|
1.612
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr6_+_34205369
|
1.599
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_-_37499708
|
1.598
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr5_+_149546340
|
1.596
|
NM_001804
|
CDX1
|
caudal type homeobox 1
|
|
chr10_-_129924467
|
1.594
|
NM_001145966
NM_002417
|
MKI67
|
antigen identified by monoclonal antibody Ki-67
|
|
chr6_+_34204663
|
1.594
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr17_-_63052816
|
1.590
|
|
|
|
|
chr15_-_93616308
|
1.588
|
|
RGMA
|
RGM domain family, member A
|
|
chr17_-_27916602
|
1.580
|
NM_001085454
NM_014030
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr1_+_41249479
|
1.574
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr15_-_101792125
|
1.572
|
NM_014918
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr1_-_200992824
|
1.571
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr1_+_43148359
|
1.570
|
|
YBX1
|
Y box binding protein 1
|
|
chr19_-_6110561
|
1.556
|
NM_000635
NM_134433
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr1_+_43148142
|
1.554
|
|
YBX1
|
Y box binding protein 1
|
|
chr14_-_91884138
|
1.546
|
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr1_-_111217625
|
1.545
|
NM_002232
|
KCNA3
|
potassium voltage-gated channel, shaker-related subfamily, member 3
|
|
chr22_-_38713384
|
1.543
|
NM_152221
|
CSNK1E
|
casein kinase 1, epsilon
|
|
chr6_+_35420149
|
1.543
|
|
FANCE
|
Fanconi anemia, complementation group E
|
|
chr11_+_66624875
|
1.536
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr18_+_42259891
|
1.532
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr17_+_4613931
|
1.527
|
|
ARRB2
|
arrestin, beta 2
|
|
chr9_-_110252045
|
1.526
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr21_+_38739858
|
1.525
|
NM_101395
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr6_-_91006580
|
1.524
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_-_43453724
|
1.524
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr14_+_105780903
|
1.514
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chrX_-_153095810
|
1.512
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr9_+_101867471
|
1.511
|
|
|
|
|
chrX_-_153095996
|
1.509
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr2_+_174219537
|
1.508
|
NM_031942
NM_145810
|
CDCA7
|
cell division cycle associated 7
|
|
chr16_+_15744082
|
1.505
|
NM_017668
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr22_-_38713217
|
1.490
|
|
CSNK1E
|
casein kinase 1, epsilon
|
|
chr1_-_111217275
|
1.489
|
|
KCNA3
|
potassium voltage-gated channel, shaker-related subfamily, member 3
|
|
chr14_+_75894919
|
1.488
|
NM_001135048
NM_130469
|
JDP2
|
Jun dimerization protein 2
|
|
chr19_+_41107220
|
1.476
|
NM_001042545
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr17_-_63052744
|
1.473
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr4_-_78740508
|
1.469
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr2_-_100720934
|
1.467
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr19_-_10679328
|
1.462
|
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr15_-_93616358
|
1.462
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr9_-_16870719
|
1.459
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr6_-_79787784
|
1.454
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chrX_-_153095746
|
1.449
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr17_-_62658311
|
1.446
|
NM_022739
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr17_-_44270255
|
1.443
|
NM_015443
|
KIAA1267
|
KIAA1267
|
|
chr10_+_105344714
|
1.440
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr6_-_112194430
|
1.437
|
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr19_+_18794487
|
1.436
|
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr4_-_109089444
|
1.433
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr17_+_4613780
|
1.429
|
NM_004313
NM_199004
|
ARRB2
|
arrestin, beta 2
|
|
chr9_+_82187469
|
1.429
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr11_+_63581031
|
1.416
|
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chrX_-_153602998
|
1.415
|
NM_001110556
NM_001456
|
FLNA
|
filamin A, alpha
|
|
chr9_-_100954883
|
1.414
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr7_+_18126543
|
1.412
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr17_-_40428311
|
1.411
|
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr18_+_77155714
|
1.408
|
NM_006162
NM_172388
NM_172390
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr12_+_54393876
|
1.405
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr12_-_129308293
|
1.403
|
NM_145648
|
SLC15A4
|
solute carrier family 15, member 4
|
|
chrX_-_153362979
|
1.403
|
|
MECP2
LOC728653
|
methyl CpG binding protein 2 (Rett syndrome)
uncharacterized LOC728653
|
|
chr6_+_34204647
|
1.402
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_-_228135587
|
1.399
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr2_-_235405692
|
1.399
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr12_-_50101127
|
1.396
|
|
FMNL3
|
formin-like 3
|
|
chr1_+_145477018
|
1.396
|
NM_153713
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr9_+_34958320
|
1.372
|
|
KIAA1045
|
KIAA1045
|
|
chr11_-_78128852
|
1.368
|
NM_080491
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr16_-_58231641
|
1.361
|
NM_001896
|
CSNK2A2
|
casein kinase 2, alpha prime polypeptide
|
|
chr3_-_13921617
|
1.358
|
NM_004625
|
WNT7A
|
wingless-type MMTV integration site family, member 7A
|
|
chr3_+_181430075
|
1.356
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr1_+_226250413
|
1.349
|
|
H3F3A
|
H3 histone, family 3A
|
|
chr11_-_46142955
|
1.348
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr2_+_28616346
|
1.337
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr8_+_95835586
|
1.330
|
|
INTS8
|
integrator complex subunit 8
|
|
chr17_-_10101854
|
1.325
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr22_-_36784055
|
1.321
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr2_-_175547471
|
1.312
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr20_-_52210780
|
1.304
|
|
ZNF217
|
zinc finger protein 217
|
|
chr8_-_101733735
|
1.294
|
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr20_-_50159247
|
1.292
|
NM_012340
NM_173091
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr1_+_29241002
|
1.292
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr9_-_134615216
|
1.289
|
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr7_-_132261268
|
1.287
|
NM_001105543
NM_020911
|
PLXNA4
|
plexin A4
|
|
chr17_-_62658175
|
1.286
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr1_+_154298034
|
1.284
|
NM_001005855
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr12_-_1703330
|
1.273
|
NM_152441
|
FBXL14
|
F-box and leucine-rich repeat protein 14
|
|
chr7_+_143078608
|
1.270
|
|
ZYX
|
zyxin
|
|
chr12_-_120554558
|
1.269
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr2_-_240322692
|
1.268
|
|
HDAC4
|
histone deacetylase 4
|
|
chr1_+_12227075
|
1.267
|
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr2_+_70314781
|
1.266
|
|
PCBP1
|
poly(rC) binding protein 1
|
|
chr4_+_38665587
|
1.264
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr1_-_3447974
|
1.262
|
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr15_+_40733302
|
1.254
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr11_-_3862137
|
1.249
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr4_-_102268626
|
1.246
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr10_-_30025754
|
1.244
|
|
SVIL
|
supervillin
|
|
chr14_+_105886253
|
1.239
|
|
MTA1
|
metastasis associated 1
|
|
chr12_-_120554557
|
1.238
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr9_-_96717544
|
1.237
|
NM_021570
|
BARX1
|
BARX homeobox 1
|
|
chr3_-_185542807
|
1.235
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr7_+_143078399
|
1.234
|
|
ZYX
|
zyxin
|
|
chr2_-_100721128
|
1.233
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr8_+_95835517
|
1.231
|
NM_017864
|
INTS8
|
integrator complex subunit 8
|
|
chr5_-_65017874
|
1.229
|
NM_019072
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr14_-_74253876
|
1.227
|
NM_001043318
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr1_+_90098693
|
1.220
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|