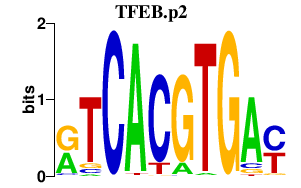
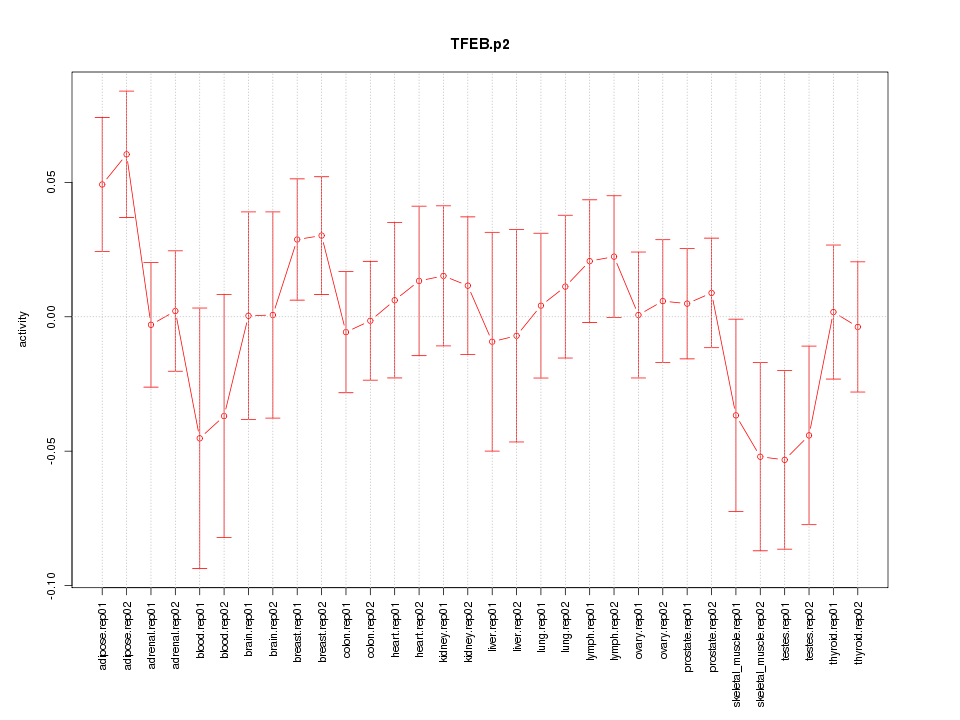
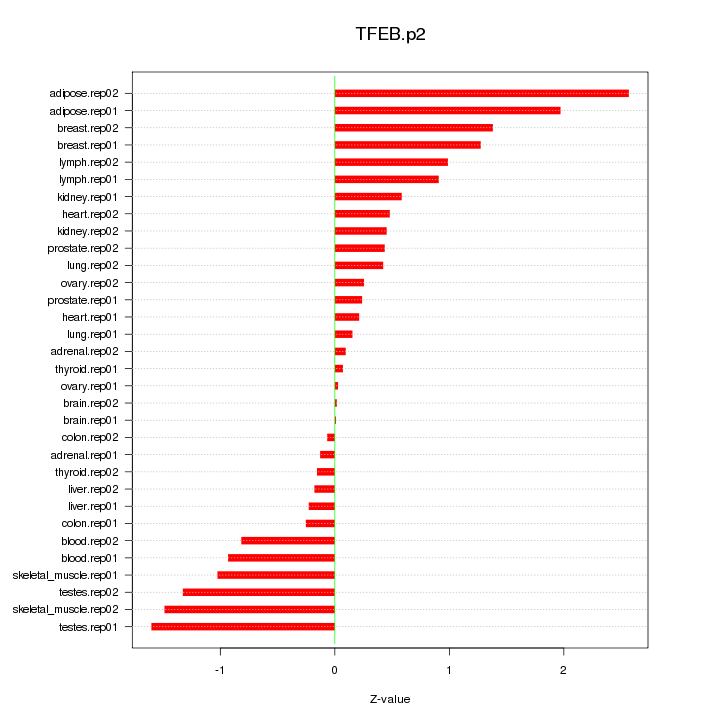
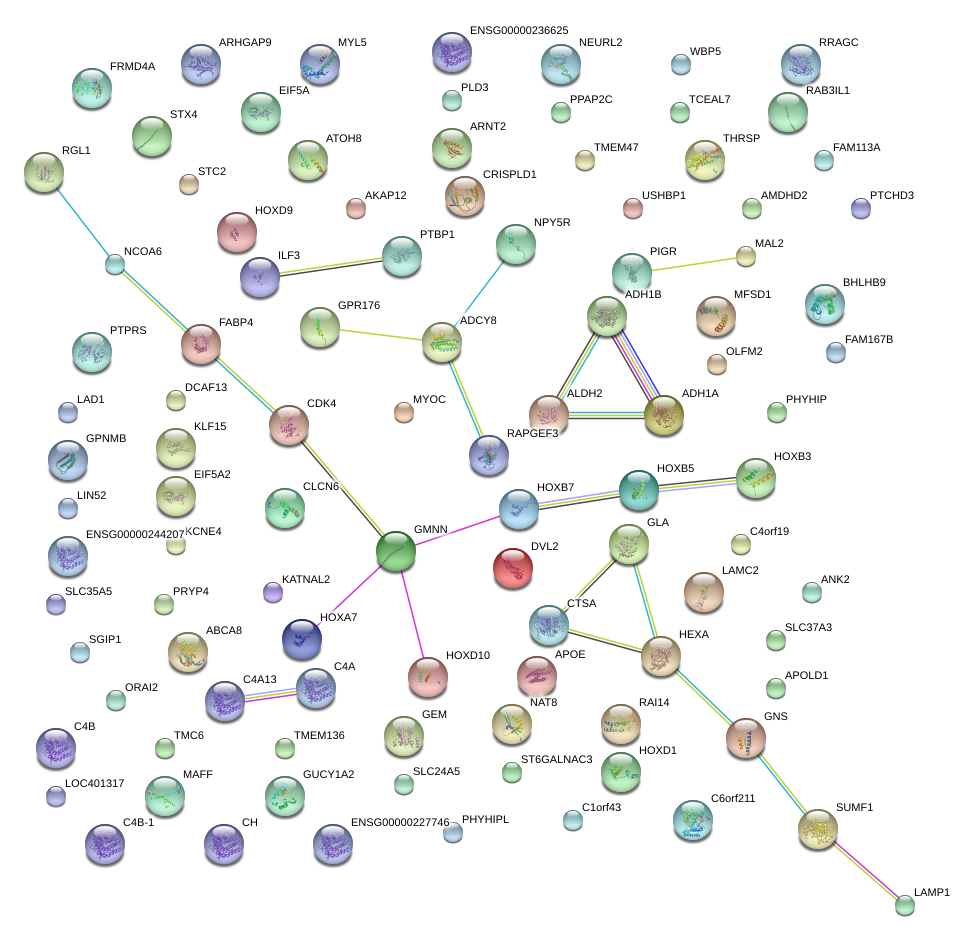
|
chr17_-_46688279
|
3.725
|
|
HOXB7
|
homeobox B7
|
|
chr8_-_82395401
|
3.644
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr17_-_46688362
|
3.547
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr4_-_100242487
|
3.084
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr10_-_14050453
|
2.826
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr19_-_10764449
|
2.774
|
|
LOC147727
|
uncharacterized LOC147727
|
|
chr13_+_113951537
|
2.726
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr20_-_44519629
|
2.691
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr7_-_27196163
|
2.625
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr2_+_176981491
|
2.484
|
NM_002148
|
HOXD10
|
homeobox D10
|
|
chr13_+_113951461
|
2.461
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr1_+_11866240
|
2.094
|
|
CLCN6
|
chloride channel 6
|
|
chr19_-_5340700
|
2.038
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr12_+_12878718
|
2.030
|
NM_001130415
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr19_+_10764896
|
2.014
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr12_-_58146108
|
1.946
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_-_154193044
|
1.850
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr17_-_66951381
|
1.846
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr1_+_11866302
|
1.800
|
|
CLCN6
|
chloride channel 6
|
|
chr5_+_34656367
|
1.778
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr12_-_58146118
|
1.766
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr3_-_4508928
|
1.748
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr4_-_100212085
|
1.712
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr12_+_54519836
|
1.708
|
|
LOC400043
|
uncharacterized LOC400043
|
|
chr15_-_40213087
|
1.689
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr7_+_23286384
|
1.670
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286390
|
1.663
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr2_+_176987412
|
1.662
|
NM_014213
|
HOXD9
|
homeobox D9
|
|
chr19_-_17375522
|
1.648
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr6_+_31982538
|
1.633
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr6_+_31949800
|
1.621
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr7_+_23286386
|
1.616
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286401
|
1.598
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr2_+_223916861
|
1.594
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr3_-_4508939
|
1.587
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr4_+_164265090
|
1.586
|
NM_006174
|
NPY5R
|
neuropeptide Y receptor Y5
|
|
chr13_+_113951589
|
1.582
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr6_+_151646634
|
1.571
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr11_-_61684981
|
1.507
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr5_+_34656482
|
1.488
|
|
RAI14
|
retinoic acid induced 14
|
|
chr12_-_58146025
|
1.486
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr8_-_95274540
|
1.485
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr12_-_58146110
|
1.485
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr6_+_151561508
|
1.483
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr8_-_95274520
|
1.466
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr3_-_4508952
|
1.462
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr17_-_46671043
|
1.460
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr5_+_34656595
|
1.458
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr8_+_120220554
|
1.433
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr1_+_76540374
|
1.421
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr16_+_2570322
|
1.394
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr1_-_207119707
|
1.390
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr7_+_28448970
|
1.388
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr17_-_46667596
|
1.385
|
|
HOXB3
|
homeobox B3
|
|
chr15_-_72668301
|
1.383
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr11_+_77774902
|
1.377
|
NM_003251
|
THRSP
|
thyroid hormone responsive
|
|
chr12_-_65153182
|
1.371
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr17_-_46687925
|
1.349
|
|
HOXB7
|
homeobox B7
|
|
chr19_+_40854529
|
1.349
|
|
PLD3
|
phospholipase D family, member 3
|
|
chrX_+_102585113
|
1.335
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr19_+_10764987
|
1.335
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr19_+_45409003
|
1.316
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr6_-_169650846
|
1.316
|
|
|
|
|
chr7_+_23286420
|
1.287
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_+_32712817
|
1.284
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chrX_-_100662905
|
1.276
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr17_-_7137722
|
1.270
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr6_+_151561094
|
1.260
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr16_+_31044849
|
1.250
|
|
STX4
|
syntaxin 4
|
|
chr12_-_48152860
|
1.232
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr18_+_44497512
|
1.224
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr3_-_4508955
|
1.220
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr4_+_37455551
|
1.220
|
NM_001104629
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chrX_-_100662788
|
1.217
|
|
GLA
|
galactosidase, alpha
|
|
chr8_+_75896707
|
1.210
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr1_-_201368434
|
1.172
|
|
LAD1
|
ladinin 1
|
|
chr19_-_10047014
|
1.150
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chrX_-_34675366
|
1.146
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr5_-_172755195
|
1.136
|
|
STC2
|
stanniocalcin 2
|
|
chr16_+_31044881
|
1.119
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr11_-_106888829
|
1.103
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr2_+_177053306
|
1.102
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr3_+_158519911
|
1.092
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr1_-_39325317
|
1.082
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr12_-_65153148
|
1.074
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr12_-_65153055
|
1.072
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr3_+_112280871
|
1.061
|
NM_017945
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr7_+_102074002
|
1.058
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr6_+_151561571
|
1.054
|
|
|
|
|
chr20_+_44520021
|
1.042
|
|
CTSA
|
cathepsin A
|
|
chr7_+_27135983
|
1.041
|
|
|
|
|
chr17_-_76124791
|
1.040
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr3_+_158519888
|
1.038
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr19_-_291335
|
1.030
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr22_+_38599026
|
1.029
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr19_+_797444
|
1.021
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr2_+_85980600
|
1.019
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr4_+_668250
|
1.019
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr3_-_170626425
|
1.011
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr10_-_27703178
|
0.996
|
NM_001034842
|
PTCHD3
|
patched domain containing 3
|
|
chr17_-_7137580
|
0.996
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr8_-_22089517
|
0.978
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr1_-_171621750
|
0.975
|
NM_000261
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response
|
|
chr20_-_2821245
|
0.973
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr3_+_112280911
|
0.965
|
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr7_+_27135672
|
0.965
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chrX_+_101975641
|
0.955
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chrX_+_102611379
|
0.954
|
NM_001006612
NM_001006613
NM_001006614
NM_016303
|
WBP5
|
WW domain binding protein 5
|
|
chr3_-_126076057
|
0.954
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr11_-_71814294
|
0.942
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr1_+_183605207
|
0.932
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr6_+_151561653
|
0.931
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr3_-_170626381
|
0.930
|
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr6_+_151773421
|
0.930
|
NM_024573
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr5_+_148786439
|
0.928
|
|
MIR143HG
|
MIR143 host gene (non-protein coding)
|
|
chr1_+_183155393
|
0.914
|
|
LAMC2
|
laminin, gamma 2
|
|
chr15_+_48413168
|
0.899
|
NM_205850
|
SLC24A5
|
solute carrier family 24, member 5
|
|
chr1_+_66999805
|
0.897
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr11_-_71814335
|
0.895
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr4_+_113739203
|
0.886
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr7_-_140098256
|
0.886
|
NM_032295
NM_207113
|
SLC37A3
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 3
|
|
chr14_+_74551655
|
0.885
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr11_+_120195837
|
0.875
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chr1_-_154192882
|
0.872
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr9_-_104249333
|
0.867
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr20_+_44519965
|
0.867
|
|
CTSA
|
cathepsin A
|
|
chr2_+_187558654
|
0.867
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr4_+_128554079
|
0.865
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr11_+_101980736
|
0.856
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr15_-_72668184
|
0.854
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr17_+_79885567
|
0.826
|
|
LOC92659
|
uncharacterized LOC92659
|
|
chr3_+_51428720
|
0.824
|
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr2_+_238395863
|
0.824
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr5_+_41904459
|
0.824
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr18_+_12407894
|
0.818
|
NM_001142405
NM_006553
|
SLMO1
|
slowmo homolog 1 (Drosophila)
|
|
chr14_-_92572950
|
0.815
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr14_-_54955664
|
0.811
|
|
GMFB
|
glia maturation factor, beta
|
|
chr11_-_71814371
|
0.808
|
NM_017907
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr12_-_59314069
|
0.807
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chrX_-_100914816
|
0.805
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr17_-_76124767
|
0.802
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr7_-_27142301
|
0.797
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr11_+_120196004
|
0.792
|
|
TMEM136
|
transmembrane protein 136
|
|
chr19_-_11688508
|
0.792
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr1_-_154193073
|
0.791
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr19_-_11688484
|
0.784
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr3_+_33318911
|
0.779
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr21_-_43430448
|
0.778
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr12_-_65153189
|
0.776
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr6_-_151773168
|
0.775
|
NM_017909
|
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae)
|
|
chr15_-_50978960
|
0.772
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr5_+_72921980
|
0.772
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr5_+_149340346
|
0.769
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr11_-_777470
|
0.768
|
NM_182612
|
PDDC1
|
Parkinson disease 7 domain containing 1
|
|
chr2_-_183903225
|
0.763
|
NM_013436
NM_205842
|
NCKAP1
|
NCK-associated protein 1
|
|
chr20_-_2821120
|
0.755
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr2_-_197036301
|
0.752
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr12_-_56122800
|
0.750
|
|
CD63
|
CD63 molecule
|
|
chr12_-_56122840
|
0.745
|
NM_001040034
NM_001780
|
CD63
|
CD63 molecule
|
|
chr8_+_26240522
|
0.744
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr11_+_9685650
|
0.741
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr12_-_63328857
|
0.740
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr7_+_100464949
|
0.737
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr1_+_199996729
|
0.735
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr11_+_9685606
|
0.732
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr5_+_149340437
|
0.731
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr2_+_208576365
|
0.728
|
|
CCNYL1
|
cyclin Y-like 1
|
|
chr17_-_7137675
|
0.725
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr12_-_123380628
|
0.723
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr7_+_150759633
|
0.722
|
NM_001199693
NM_001199694
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr12_-_56122792
|
0.715
|
|
CD63
|
CD63 molecule
|
|
chr11_-_85780105
|
0.710
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_-_56122748
|
0.706
|
|
CD63
|
CD63 molecule
|
|
chr15_+_44084493
|
0.705
|
|
SERF2
|
small EDRK-rich factor 2
|
|
chr7_-_47622127
|
0.701
|
|
TNS3
|
tensin 3
|
|
chr6_-_99873129
|
0.698
|
NM_015491
NM_032870
|
PNISR
|
PNN-interacting serine/arginine-rich protein
|
|
chr1_+_11866206
|
0.694
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr19_+_49588399
|
0.689
|
NM_003089
|
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1)
|
|
chrX_+_102631408
|
0.682
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr11_-_71814268
|
0.676
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr12_-_59314297
|
0.674
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr2_+_26987122
|
0.672
|
NM_017877
|
CENPA
C2orf18
|
centromere protein A
chromosome 2 open reading frame 18
|
|
chr7_-_27183225
|
0.671
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr8_+_98788014
|
0.668
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr6_+_42018264
|
0.666
|
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr7_-_47621741
|
0.665
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr19_+_49458031
|
0.661
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr2_+_173292306
|
0.660
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr14_+_38677186
|
0.656
|
NM_001049
|
SSTR1
|
somatostatin receptor 1
|
|
chr14_-_54955714
|
0.655
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr8_-_11725509
|
0.655
|
|
CTSB
|
cathepsin B
|
|
chr4_+_2061238
|
0.649
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr3_+_106959515
|
0.648
|
|
LOC344595
|
uncharacterized LOC344595
|
|
chr3_+_113465910
|
0.646
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr15_-_50978983
|
0.643
|
NM_017672
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr20_-_62436855
|
0.638
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr12_+_93964801
|
0.638
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr6_-_33386064
|
0.632
|
NM_001014433
NM_001014837
NM_001014838
NM_001014840
NM_015921
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr5_+_80256457
|
0.628
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr17_-_46655544
|
0.627
|
NM_024015
|
HOXB4
|
homeobox B4
|