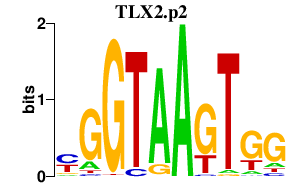
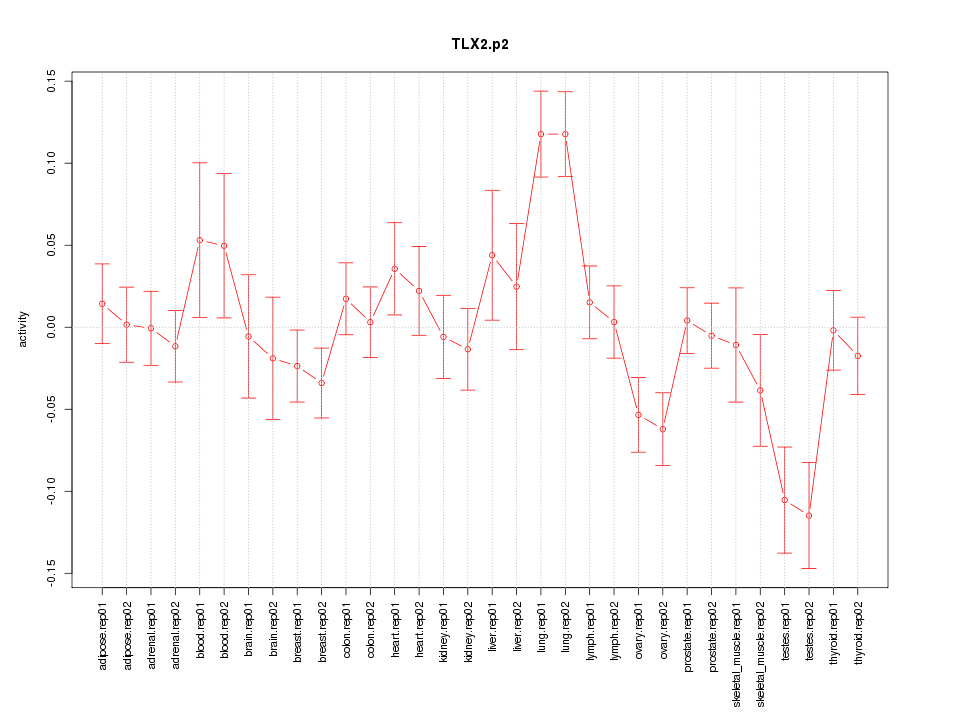
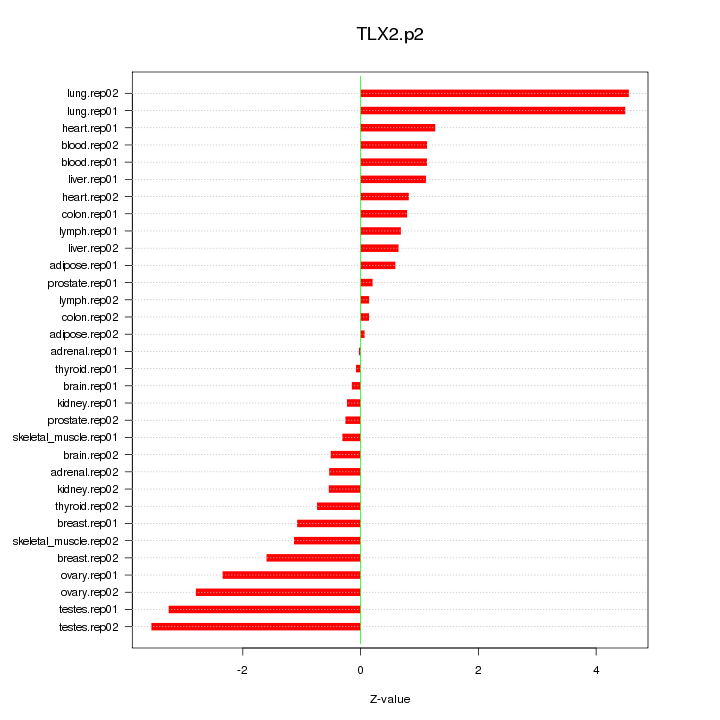
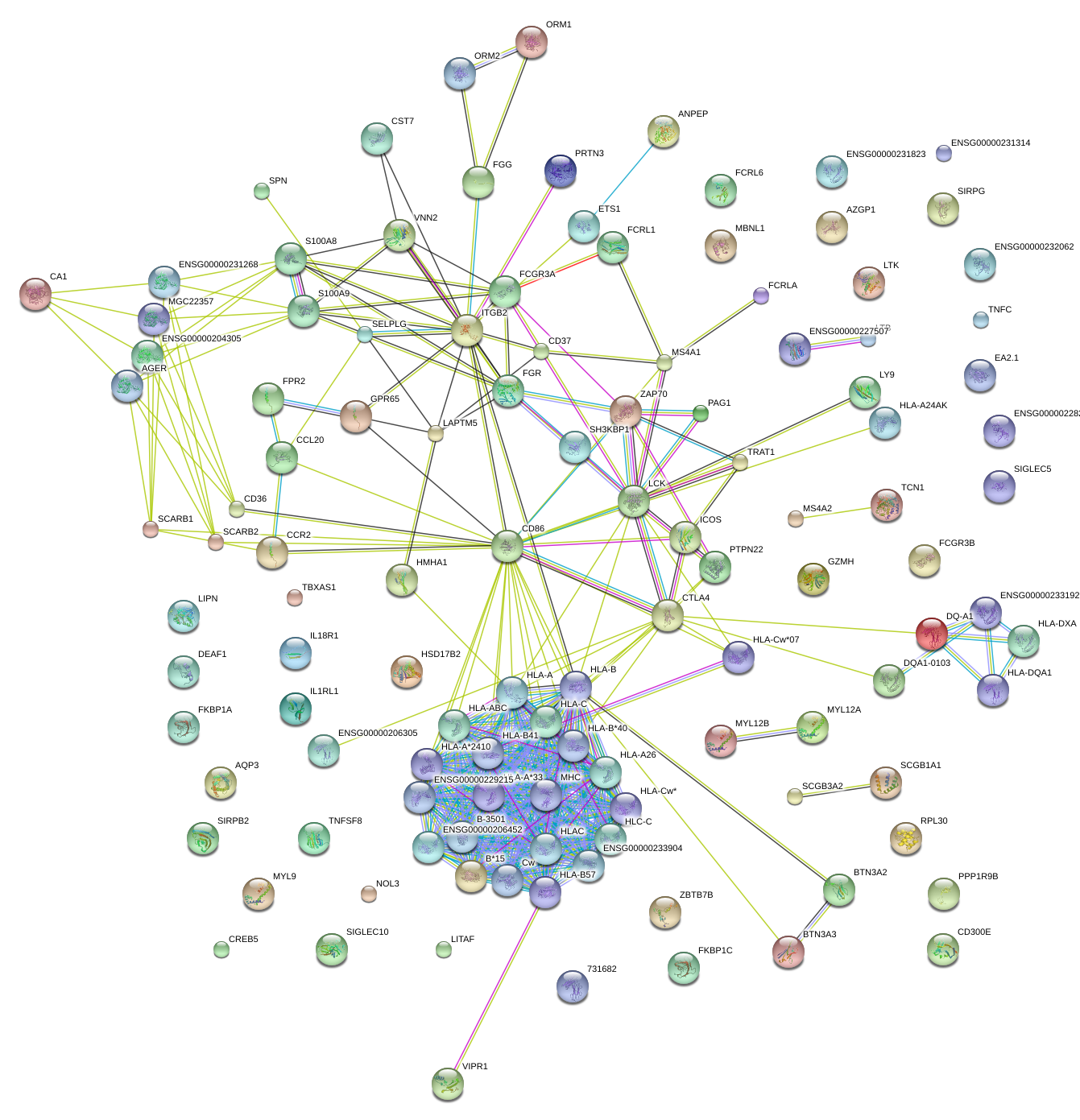
|
chr19_+_52264452
|
4.165
|
NM_001005738
|
FPR2
|
formyl peptide receptor 2
|
|
chr14_-_106321626
|
3.231
|
|
|
|
|
chr11_+_62186506
|
3.227
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr4_-_155533800
|
3.005
|
|
FGG
|
fibrinogen gamma chain
|
|
chr1_-_153363544
|
2.863
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr4_-_155533757
|
2.791
|
|
FGG
|
fibrinogen gamma chain
|
|
chr11_+_60223281
|
2.478
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr9_+_117085302
|
2.389
|
NM_000607
|
ORM1
|
orosomucoid 1
|
|
chr3_+_42544094
|
2.308
|
NM_001251883
NM_001251884
NM_001251885
NM_004624
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr1_+_160765927
|
2.280
|
NM_001033667
NM_002348
|
LY9
|
lymphocyte antigen 9
|
|
chr9_+_117092068
|
2.264
|
NM_000608
|
ORM2
|
orosomucoid 2
|
|
chr4_-_155533850
|
2.249
|
|
FGG
|
fibrinogen gamma chain
|
|
chr2_+_102927961
|
2.236
|
NM_016232
|
IL18R1
IL1RL1
|
interleukin 18 receptor 1
interleukin 1 receptor-like 1
|
|
chr3_+_42544131
|
2.203
|
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr4_-_155533882
|
2.148
|
NM_000509
NM_021870
|
FGG
|
fibrinogen gamma chain
|
|
chr2_+_102979096
|
1.990
|
NM_003855
|
IL18R1
|
interleukin 18 receptor 1
|
|
chr6_-_32151996
|
1.974
|
NM_001136
NM_001206929
NM_001206932
NM_001206934
NM_001206936
NM_001206940
NM_001206954
NM_001206966
NM_172197
|
AGER
|
advanced glycosylation end product-specific receptor
|
|
chr1_+_153330329
|
1.969
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr5_+_147258273
|
1.952
|
NM_054023
|
SCGB3A2
|
secretoglobin, family 3A, member 2
|
|
chr15_-_41805993
|
1.871
|
NM_001135685
NM_002344
NM_206961
|
LTK
|
leukocyte receptor tyrosine kinase
|
|
chr17_-_72619794
|
1.860
|
NM_181449
|
CD300E
|
CD300e molecule
|
|
chr1_+_159772172
|
1.858
|
NM_001004310
|
FCRL6
|
Fc receptor-like 6
|
|
chr6_-_133084585
|
1.858
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr14_-_25078818
|
1.841
|
NM_033423
|
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX)
|
|
chr11_-_59633942
|
1.718
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr8_-_82024277
|
1.713
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr8_-_99057594
|
1.709
|
|
RPL30
|
ribosomal protein L30
|
|
chr15_-_90347534
|
1.709
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr3_+_121774208
|
1.706
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chr1_-_114414245
|
1.694
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr1_+_32739711
|
1.692
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr9_-_33447527
|
1.686
|
NM_004925
|
AQP3
|
aquaporin 3 (Gill blood group)
|
|
chr8_-_99057751
|
1.679
|
|
RPL30
|
ribosomal protein L30
|
|
chr6_+_32605182
|
1.675
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr2_+_228678556
|
1.657
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr15_-_31523049
|
1.656
|
NM_001243538
|
LOC283710
|
uncharacterized LOC283710
|
|
chr2_+_204801453
|
1.656
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr2_+_98330030
|
1.583
|
NM_001079
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chr20_+_24929904
|
1.582
|
|
CST7
|
cystatin F (leukocystatin)
|
|
chr3_+_108541544
|
1.568
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr17_+_57915192
|
1.564
|
|
|
|
|
chr1_-_161601251
|
1.558
|
NM_001244753
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b)
|
|
chr7_+_28475233
|
1.541
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr16_+_82068910
|
1.522
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr1_-_31230594
|
1.513
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chrX_-_19817854
|
1.504
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr6_-_31550065
|
1.502
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr8_-_99057766
|
1.489
|
NM_000989
|
RPL30
|
ribosomal protein L30
|
|
chr1_-_27953046
|
1.489
|
NM_001042729
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr19_+_840970
|
1.473
|
NM_002777
|
PRTN3
|
proteinase 3
|
|
chr16_+_82068958
|
1.464
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr12_-_109027565
|
1.464
|
|
SELPLG
|
selectin P ligand
|
|
chr1_-_161519526
|
1.463
|
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr19_-_51920841
|
1.441
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr7_+_139528951
|
1.439
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr19_+_49838700
|
1.437
|
|
CD37
|
CD37 molecule
|
|
chr7_-_99573637
|
1.436
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr16_+_29674568
|
1.432
|
NM_003123
|
SPN
|
sialophorin
|
|
chr19_+_1067293
|
1.427
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr19_-_52150007
|
1.401
|
NM_001098612
|
SIGLEC14
|
sialic acid binding Ig-like lectin 14
|
|
chr11_-_128391888
|
1.400
|
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr20_-_1373744
|
1.394
|
NM_000801
NM_001199786
NM_054014
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr20_-_1472004
|
1.391
|
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr16_+_29674979
|
1.381
|
|
SPN
|
sialophorin
|
|
chrX_-_19817907
|
1.375
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_-_157789832
|
1.360
|
NM_001159397
NM_001159398
NM_052938
|
FCRL1
|
Fc receptor-like 1
|
|
chr12_-_109027654
|
1.347
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr18_+_3247774
|
1.342
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr12_-_109027615
|
1.339
|
|
SELPLG
|
selectin P ligand
|
|
chr16_+_82068830
|
1.337
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr7_+_80267972
|
1.322
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr10_+_90521162
|
1.320
|
NM_001102469
|
LIPN
|
lipase, family member N
|
|
chr8_-_86290333
|
1.304
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr20_-_1373702
|
1.302
|
|
FKBP1A-SDCBP2
FKBP1A
|
FKBP1A-SDCBP2 readthrough (non-protein coding)
FK506 binding protein 1A, 12kDa
|
|
chr3_+_151986731
|
1.300
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr14_+_88471467
|
1.298
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr19_-_52133589
|
1.293
|
NM_003830
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5
|
|
chr1_+_154975041
|
1.288
|
NM_001252405
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr9_-_117692874
|
1.283
|
NM_001244
NM_001252290
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8
|
|
chr21_-_46340776
|
1.272
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr19_+_49838646
|
1.271
|
|
CD37
|
CD37 molecule
|
|
chr6_+_26365397
|
1.269
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr3_+_46395574
|
1.269
|
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr16_+_29674564
|
1.263
|
|
SPN
|
sialophorin
|
|
chr16_-_11680756
|
1.261
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr6_-_31324955
|
1.253
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr1_-_153518265
|
1.251
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr20_-_1472168
|
1.226
|
NM_001122962
NM_001134836
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr1_-_153348057
|
1.225
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr19_+_7733971
|
1.218
|
NM_001193374
NM_020415
|
RETN
|
resistin
|
|
chr3_+_46395224
|
1.215
|
NM_001123041
NM_001123396
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr5_-_157002739
|
1.211
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr3_-_112693711
|
1.211
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr6_-_32557431
|
1.210
|
NM_001243965
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr20_-_35274557
|
1.204
|
NM_032214
NM_175077
|
SLA2
|
Src-like-adaptor 2
|
|
chr19_+_49838673
|
1.202
|
NM_001040031
NM_001774
|
CD37
|
CD37 molecule
|
|
chr7_+_48128939
|
1.200
|
|
UPP1
|
uridine phosphorylase 1
|
|
chr7_-_38375172
|
1.199
|
|
TRGV7
|
T cell receptor gamma variable 7 (pseudogene)
|
|
chr12_-_51740422
|
1.197
|
NM_001971
|
CELA1
|
chymotrypsin-like elastase family, member 1
|
|
chr19_-_8642337
|
1.193
|
|
MYO1F
|
myosin IF
|
|
chr3_-_46249787
|
1.192
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr1_+_154975201
|
1.181
|
NM_001252406
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr3_-_182880463
|
1.178
|
NM_014398
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr21_-_46340821
|
1.178
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr3_+_121796695
|
1.177
|
NM_006889
NM_176892
|
CD86
|
CD86 molecule
|
|
chr1_+_111772332
|
1.177
|
NM_001025199
|
CHI3L2
|
chitinase 3-like 2
|
|
chr10_-_135090325
|
1.177
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr6_-_39282095
|
1.175
|
NM_031460
NM_001135111
|
KCNK17
|
potassium channel, subfamily K, member 17
|
|
chr20_+_37434294
|
1.172
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr4_-_145061787
|
1.171
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chr18_+_3247825
|
1.163
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr14_-_106110894
|
1.158
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr20_-_1472126
|
1.155
|
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr19_+_16999871
|
1.155
|
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr19_-_8642232
|
1.151
|
|
MYO1F
|
myosin IF
|
|
chr1_-_157746863
|
1.145
|
NM_030764
|
FCRL2
|
Fc receptor-like 2
|
|
chr1_-_25291474
|
1.143
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr17_+_7239847
|
1.141
|
NM_014716
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|
|
chr19_+_827821
|
1.135
|
NM_001700
|
AZU1
|
azurocidin 1
|
|
chr20_+_54987167
|
1.131
|
NM_001164116
NM_001164114
NM_001164115
NM_020356
|
CASS4
|
Cas scaffolding protein family member 4
|
|
chr1_-_160990862
|
1.125
|
|
F11R
|
F11 receptor
|
|
chr20_+_24929862
|
1.117
|
NM_003650
|
CST7
|
cystatin F (leukocystatin)
|
|
chr14_-_106725620
|
1.111
|
|
IGHV4-31
IGHV3-23
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-23
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr19_-_8642309
|
1.111
|
NM_012335
|
MYO1F
|
myosin IF
|
|
chr6_-_32497925
|
1.109
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr5_-_39270708
|
1.109
|
NM_001243093
|
FYB
|
FYN binding protein
|
|
chr6_+_31554975
|
1.095
|
NM_001166538
NM_007161
NM_205840
|
LST1
|
leukocyte specific transcript 1
|
|
chr19_+_1026271
|
1.095
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr16_+_81478774
|
1.093
|
NM_198390
|
CMIP
|
c-Maf inducing protein
|
|
chr6_-_31324975
|
1.088
|
NM_005514
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr6_-_32920812
|
1.088
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr20_-_62710787
|
1.087
|
NM_005873
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr7_+_99971067
|
1.085
|
NM_013439
NM_178272
NM_178273
|
PILRA
|
paired immunoglobin-like type 2 receptor alpha
|
|
chr20_-_1373571
|
1.082
|
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chrX_+_67867510
|
1.079
|
NM_001142503
NM_014725
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr19_-_54850385
|
1.079
|
NM_012276
|
LILRA4
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4
|
|
chr18_+_61582744
|
1.079
|
NM_005024
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr19_+_840963
|
1.078
|
|
PRTN3
|
proteinase 3
|
|
chr18_+_3247815
|
1.068
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr3_+_151531860
|
1.062
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr1_-_161519817
|
1.056
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr7_+_99971132
|
1.037
|
|
PILRA
|
paired immunoglobin-like type 2 receptor alpha
|
|
chr6_-_31324898
|
1.035
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr16_-_89007605
|
1.034
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr19_+_1026624
|
1.032
|
|
CNN2
|
calponin 2
|
|
chr7_-_99277518
|
1.027
|
NM_000777
NM_001190484
|
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5
|
|
chr3_+_148847370
|
1.026
|
NM_032383
|
HPS3
|
Hermansky-Pudlak syndrome 3
|
|
chr4_-_38858407
|
1.022
|
NM_006068
|
TLR6
|
toll-like receptor 6
|
|
chr20_+_42544781
|
1.020
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr19_+_51728319
|
1.018
|
NM_001082618
NM_001177608
NM_001772
|
CD33
|
CD33 molecule
|
|
chr1_+_207495070
|
1.017
|
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr22_+_43547558
|
1.016
|
|
TSPO
|
translocator protein (18kDa)
|
|
chr13_+_24553919
|
1.014
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr17_-_7017558
|
1.013
|
NM_080914
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr19_+_1026627
|
1.011
|
|
CNN2
|
calponin 2
|
|
chr1_+_207495086
|
1.010
|
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr19_+_859658
|
1.007
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr7_+_139478033
|
1.002
|
NM_001130966
NM_001166254
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr4_+_166248816
|
1.002
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr2_+_169923531
|
1.001
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr6_+_31371355
|
1.000
|
NM_000247
NM_001177519
|
MICA
|
MHC class I polypeptide-related sequence A
|
|
chr1_+_26856259
|
1.000
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr18_-_28681885
|
0.999
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr6_+_106546736
|
0.999
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr19_+_1026608
|
0.998
|
|
CNN2
|
calponin 2
|
|
chr1_+_207495009
|
0.994
|
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr10_+_11865310
|
0.988
|
NM_153256
|
C10orf47
|
chromosome 10 open reading frame 47
|
|
chr11_-_118084073
|
0.977
|
NM_153206
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr4_+_74270832
|
0.976
|
|
ALB
|
albumin
|
|
chr1_+_26872306
|
0.975
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chrX_+_70765479
|
0.972
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr19_-_11688508
|
0.970
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr18_+_52495618
|
0.969
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr13_+_97999289
|
0.968
|
|
|
|
|
chr1_-_169599349
|
0.966
|
NM_003005
|
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62)
|
|
chr1_+_161551128
|
0.964
|
NM_201563
|
FCGR2C
|
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr3_+_111261039
|
0.959
|
|
CD96
|
CD96 molecule
|
|
chr7_+_80267922
|
0.958
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr11_+_65082288
|
0.957
|
NM_006779
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chrX_+_78200828
|
0.952
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr22_-_39639967
|
0.940
|
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr7_+_150434679
|
0.939
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr3_+_157154579
|
0.938
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr6_+_32407618
|
0.936
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr12_+_13044352
|
0.934
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr15_-_38856830
|
0.933
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr1_-_160990971
|
0.927
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr4_-_83347222
|
0.925
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr19_+_51728369
|
0.925
|
|
CD33
|
CD33 molecule
|
|
chr8_+_86376130
|
0.924
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr19_-_8642297
|
0.922
|
|
MYO1F
|
myosin IF
|
|
chr2_+_63815742
|
0.922
|
NM_005917
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr19_+_16999807
|
0.913
|
NM_003950
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr22_+_24823529
|
0.911
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr1_+_16174358
|
0.909
|
NM_015001
|
SPEN
|
spen homolog, transcriptional regulator (Drosophila)
|
|
chr1_+_207494816
|
0.906
|
NM_000574
NM_001114752
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr11_-_128457452
|
0.905
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_+_24286311
|
0.903
|
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr1_+_165600433
|
0.902
|
|
MGST3
|
microsomal glutathione S-transferase 3
|
|
chr8_-_134584012
|
0.900
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|