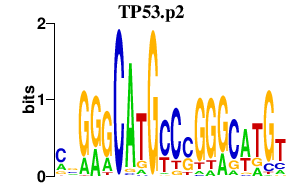
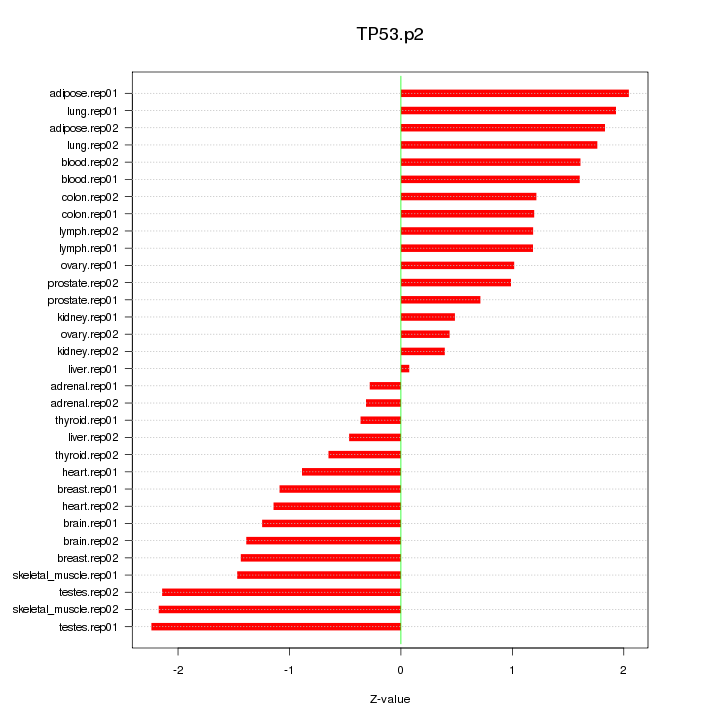
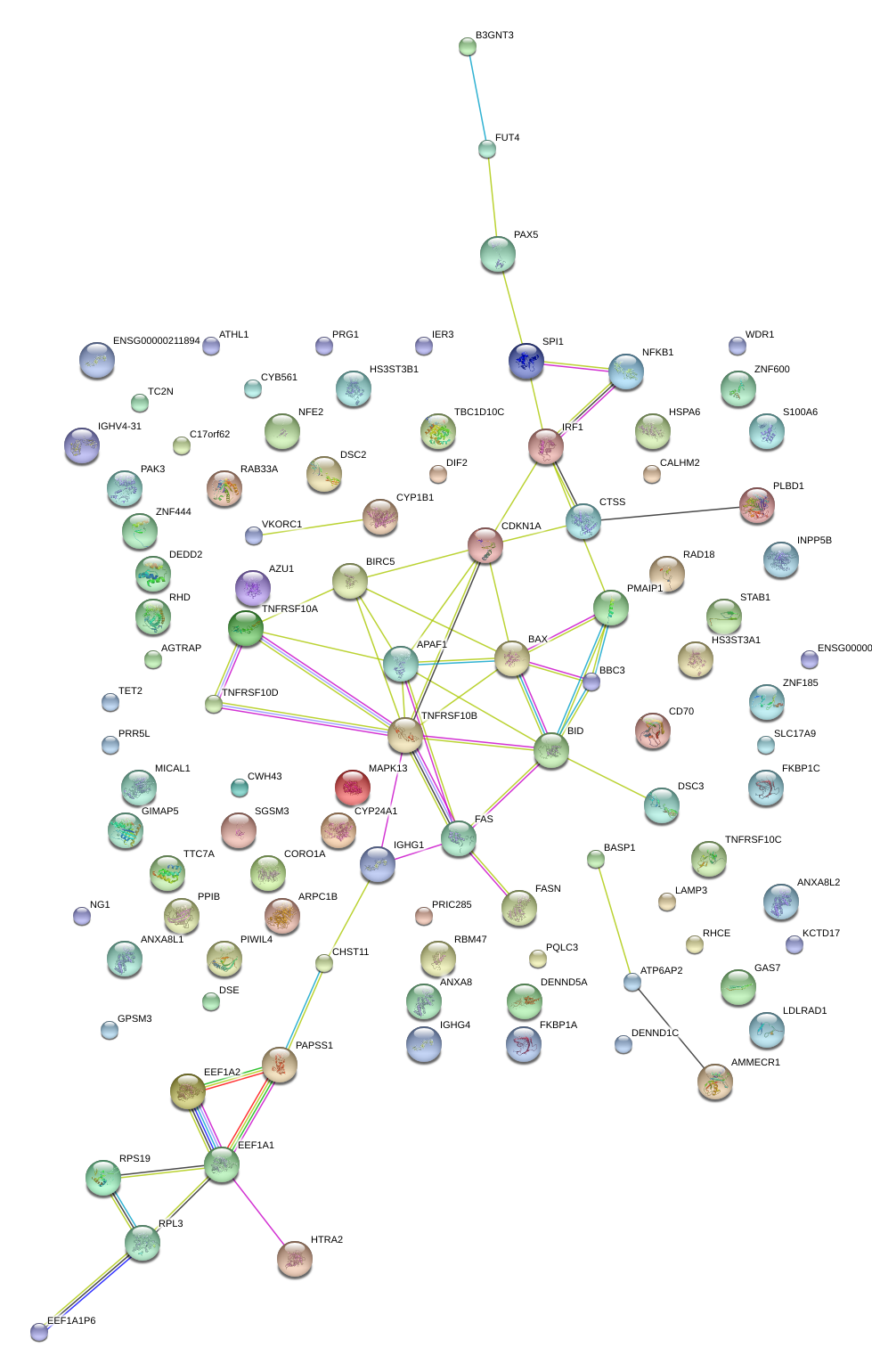
|
chr8_+_22960326
|
4.490
|
NM_003841
|
TNFRSF10C
|
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain
|
|
chr11_-_47400096
|
3.725
|
NM_001080547
NM_003120
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr11_-_47399986
|
3.084
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr11_-_47399903
|
3.038
|
|
|
|
|
chr19_+_827821
|
2.840
|
NM_001700
|
AZU1
|
azurocidin 1
|
|
chr8_-_23021447
|
2.818
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr8_-_23082572
|
2.817
|
|
TNFRSF10A
|
tumor necrosis factor receptor superfamily, member 10a
|
|
chr18_+_57567191
|
2.643
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr10_-_105212149
|
2.514
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr10_-_105212022
|
2.497
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr22_-_18257194
|
2.420
|
|
BID
|
BH3 interacting domain death agonist
|
|
chr17_+_76210329
|
2.310
|
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr8_-_22926406
|
2.248
|
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr11_+_36397534
|
2.126
|
NM_001160168
NM_024841
|
PRR5L
|
proline rich 5 like
|
|
chr11_+_289113
|
2.096
|
NM_025092
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast)
|
|
chr10_+_90751044
|
2.046
|
|
FAS
|
Fas (TNF receptor superfamily, member 6)
|
|
chr6_-_32163299
|
2.026
|
NM_022107
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr6_+_36098261
|
1.989
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr1_-_150738232
|
1.977
|
|
CTSS
|
cathepsin S
|
|
chr14_-_106610338
|
1.967
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_+_11796211
|
1.850
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr19_-_6591082
|
1.840
|
|
CD70
|
CD70 molecule
|
|
chr4_-_10118358
|
1.822
|
|
WDR1
|
WD repeat domain 1
|
|
chr1_+_11796177
|
1.711
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr2_+_11295602
|
1.683
|
|
PQLC3
|
PQ loop repeat containing 3
|
|
chr2_+_11295569
|
1.584
|
|
PQLC3
|
PQ loop repeat containing 3
|
|
chr12_-_14720790
|
1.577
|
NM_024829
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr15_-_64455206
|
1.480
|
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr4_-_40516567
|
1.476
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_-_38412725
|
1.463
|
NM_005540
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr2_+_11295539
|
1.458
|
NM_152391
|
PQLC3
|
PQ loop repeat containing 3
|
|
chr22_-_18257221
|
1.449
|
|
BID
|
BH3 interacting domain death agonist
|
|
chr11_+_94277566
|
1.433
|
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr4_-_10118378
|
1.426
|
|
WDR1
|
WD repeat domain 1
|
|
chr6_-_109775423
|
1.423
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr19_+_17905935
|
1.409
|
|
B3GNT3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3
|
|
chr10_+_90750384
|
1.368
|
|
FAS
|
Fas (TNF receptor superfamily, member 6)
|
|
chr1_+_11796128
|
1.368
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr8_-_22926490
|
1.358
|
NM_003842
NM_147187
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr2_+_47168362
|
1.332
|
|
TTC7A
|
tetratricopeptide repeat domain 7A
|
|
chr1_-_150738292
|
1.318
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr16_-_31106125
|
1.310
|
NM_024006
NM_206824
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr11_+_67171379
|
1.300
|
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr15_-_64455158
|
1.277
|
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr15_-_64455184
|
1.254
|
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr16_+_30194730
|
1.238
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr5_+_17217474
|
1.217
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr15_-_64455299
|
1.200
|
NM_000942
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr12_-_14720553
|
1.199
|
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr6_-_74230732
|
1.197
|
NM_001402
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr1_-_153508495
|
1.195
|
|
S100A6
|
S100 calcium binding protein A6
|
|
chr6_+_116692098
|
1.190
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr20_-_1373571
|
1.188
|
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr1_-_38412720
|
1.176
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr4_+_106067031
|
1.175
|
NM_017628
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr12_+_99038918
|
1.153
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr20_-_1373702
|
1.152
|
|
FKBP1A-SDCBP2
FKBP1A
|
FKBP1A-SDCBP2 readthrough (non-protein coding)
FK506 binding protein 1A, 12kDa
|
|
chr19_-_42721802
|
1.147
|
NM_133328
|
DEDD2
|
death effector domain containing 2
|
|
chr2_-_38303265
|
1.134
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chrX_+_129305553
|
1.122
|
NM_004794
|
RAB33A
|
RAB33A, member RAS oncogene family
|
|
chr2_-_38303307
|
1.119
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr1_-_150738255
|
1.114
|
|
CTSS
|
cathepsin S
|
|
chr15_-_64455190
|
1.102
|
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr2_+_74757127
|
1.100
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr16_-_31106093
|
1.096
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr22_-_39715609
|
1.092
|
NM_000967
NM_001033853
|
RPL3
|
ribosomal protein L3
|
|
chr4_-_108641291
|
1.091
|
NM_005443
|
PAPSS1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1
|
|
chr4_-_10118286
|
1.090
|
|
|
|
|
chr4_+_103422479
|
1.087
|
NM_001165412
NM_003998
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr7_+_150434679
|
1.085
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr19_+_42364322
|
1.071
|
|
RPS19
|
ribosomal protein S19
|
|
chr7_+_98972351
|
1.069
|
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa
|
|
chr6_+_36644236
|
1.050
|
NM_001220777
NM_078467
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr16_-_31106049
|
1.048
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr18_-_28622624
|
1.032
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr19_-_6481765
|
1.023
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr20_-_62205559
|
1.019
|
NM_001037335
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr20_+_61583998
|
1.017
|
NM_022082
|
SLC17A9
|
solute carrier family 17, member 9
|
|
chr2_+_74757116
|
1.017
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr19_+_49458129
|
1.012
|
|
BAX
|
BCL2-associated X protein
|
|
chr3_+_127771342
|
1.006
|
|
|
|
|
chrX_+_152086408
|
1.003
|
NM_001178113
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chrX_+_40440229
|
1.003
|
|
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2
|
|
chr4_-_10118296
|
0.996
|
|
WDR1
|
WD repeat domain 1
|
|
chr1_-_153508463
|
0.994
|
|
S100A6
|
S100 calcium binding protein A6
|
|
chr1_+_161494035
|
0.991
|
NM_002155
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B')
|
|
chr1_+_25598883
|
0.989
|
NM_001127691
NM_016124
|
RHD
|
Rh blood group, D antigen
|
|
chr1_-_54483802
|
0.988
|
NM_001010978
|
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1
|
|
chr19_-_53290015
|
0.985
|
NM_198457
|
ZNF600
|
zinc finger protein 600
|
|
chr11_+_67171400
|
0.981
|
NM_198517
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr19_-_47735858
|
0.978
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chrX_+_40440261
|
0.975
|
|
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2
|
|
chr2_+_74757158
|
0.964
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr3_-_182880432
|
0.950
|
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr17_-_61523544
|
0.947
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr19_+_42364262
|
0.942
|
|
RPS19
|
ribosomal protein S19
|
|
chr7_+_98972326
|
0.939
|
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa
|
|
chr19_+_56652689
|
0.937
|
|
ZNF444
|
zinc finger protein 444
|
|
chr22_+_40766649
|
0.930
|
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr11_-_9286832
|
0.928
|
NM_001243254
NM_015213
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr4_+_48988264
|
0.923
|
NM_025087
|
CWH43
|
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae)
|
|
chr22_+_40766563
|
0.921
|
NM_015705
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr4_-_10118444
|
0.920
|
|
WDR1
|
WD repeat domain 1
|
|
chr2_+_74756986
|
0.912
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr3_+_52529370
|
0.906
|
|
STAB1
|
stabilin 1
|
|
chr11_+_94277005
|
0.904
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr10_+_47746919
|
0.901
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr19_+_49458151
|
0.895
|
|
BAX
|
BCL2-associated X protein
|
|
chr17_+_14204326
|
0.881
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr17_-_9929567
|
0.879
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr20_-_52790511
|
0.877
|
NM_000782
NM_001128915
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1
|
|
chr6_-_30712294
|
0.873
|
NM_003897
|
IER3
|
immediate early response 3
|
|
chr17_-_80407601
|
0.867
|
NM_001193655
|
C17orf62
|
chromosome 17 open reading frame 62
|
|
chr22_+_37447615
|
0.863
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr12_-_54694647
|
0.863
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr12_+_104850986
|
0.853
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr5_-_131826426
|
0.851
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr22_+_40766636
|
0.851
|
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr20_-_1373744
|
0.847
|
NM_000801
NM_001199786
NM_054014
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr3_-_9005070
|
0.842
|
NM_020165
|
RAD18
|
RAD18 homolog (S. cerevisiae)
|
|
chr22_+_40766617
|
0.817
|
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr1_-_38412667
|
0.809
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr14_-_92302612
|
0.806
|
NM_001128595
NM_152332
|
TC2N
|
tandem C2 domains, nuclear
|
|
chrX_-_109561314
|
0.800
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr3_+_127771287
|
0.799
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr3_+_196466724
|
0.796
|
NM_002577
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr9_-_116037823
|
0.790
|
NM_139286
|
CDC26
|
cell division cycle 26 homolog (S. cerevisiae)
|
|
chr22_+_40766630
|
0.789
|
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr1_-_38412704
|
0.785
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr7_+_48128225
|
0.776
|
NM_181597
|
UPP1
|
uridine phosphorylase 1
|
|
chr4_+_103422553
|
0.774
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr1_+_151584665
|
0.767
|
|
SNX27
|
sorting nexin family member 27
|
|
chr12_+_10124007
|
0.767
|
NM_138337
NM_201623
|
CLEC12A
CLEC12B
|
C-type lectin domain family 12, member A
C-type lectin domain family 12, member B
|
|
chr16_+_57653909
|
0.764
|
NM_001145770
NM_005682
|
GPR56
|
G protein-coupled receptor 56
|
|
chr17_+_78234626
|
0.762
|
NM_020914
NM_020954
|
RNF213
|
ring finger protein 213
|
|
chrX_-_109561427
|
0.761
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr17_+_76210276
|
0.760
|
NM_001012270
NM_001012271
NM_001168
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr12_-_45270071
|
0.756
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr3_-_182880463
|
0.749
|
NM_014398
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr11_+_65154040
|
0.749
|
NM_031904
|
FRMD8
|
FERM domain containing 8
|
|
chr12_-_113772900
|
0.746
|
NM_024959
|
SLC24A6
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 6
|
|
chr21_-_44846927
|
0.745
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr16_-_31105922
|
0.740
|
|
|
|
|
chr1_-_93646245
|
0.736
|
NM_001167830
NM_016040
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr12_+_104850750
|
0.720
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr1_-_45805606
|
0.718
|
NM_001048174
NM_001048172
NM_001048173
NM_001048171
NM_001128425
NM_012222
|
MUTYH
|
mutY homolog (E. coli)
|
|
chr19_+_49458031
|
0.715
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr1_-_160832466
|
0.713
|
NM_001166663
NM_001166664
NM_016382
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4
|
|
chr1_-_93645903
|
0.710
|
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr7_+_43622638
|
0.710
|
NM_004760
|
STK17A
|
serine/threonine kinase 17a
|
|
chr19_+_17970670
|
0.708
|
NM_000980
|
RPL18A
|
ribosomal protein L18a
|
|
chr10_-_73611050
|
0.708
|
NM_001042465
NM_001042466
NM_002778
|
PSAP
|
prosaposin
|
|
chr11_+_65154093
|
0.700
|
|
FRMD8
|
FERM domain containing 8
|
|
chr21_+_30671736
|
0.697
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr2_+_74757243
|
0.694
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr15_-_50405512
|
0.692
|
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr2_+_163200623
|
0.690
|
|
GCA
|
grancalcin, EF-hand calcium binding protein
|
|
chr2_-_24307179
|
0.684
|
NM_001206802
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr12_+_104850641
|
0.683
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_+_104850784
|
0.683
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr1_-_223316589
|
0.682
|
NM_003268
|
TLR5
|
toll-like receptor 5
|
|
chr1_+_224544590
|
0.679
|
NM_014184
|
CNIH4
|
cornichon homolog 4 (Drosophila)
|
|
chr15_+_41056243
|
0.679
|
NM_005258
|
GCHFR
|
GTP cyclohydrolase I feedback regulator
|
|
chrX_+_40440145
|
0.671
|
NM_005765
|
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2
|
|
chr4_+_153457407
|
0.669
|
|
DKFZP434I0714
|
uncharacterized protein DKFZP434I0714
|
|
chr19_-_40931880
|
0.666
|
NM_013376
|
SERTAD1
|
SERTA domain containing 1
|
|
chr12_-_6451156
|
0.660
|
NM_001065
|
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A
|
|
chr2_+_113403433
|
0.659
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr19_+_5904842
|
0.654
|
NM_001017921
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein
|
|
chr5_+_135364583
|
0.652
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr20_+_61584094
|
0.648
|
|
SLC17A9
|
solute carrier family 17, member 9
|
|
chr14_+_64971226
|
0.643
|
NM_001123329
NM_014950
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr22_+_21336575
|
0.639
|
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr3_+_127771126
|
0.638
|
NM_013336
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr16_-_15149827
|
0.625
|
NM_173474
|
NTAN1
|
N-terminal asparagine amidase
|
|
chr6_+_32939371
|
0.623
|
NM_001199455
|
BRD2
|
bromodomain containing 2
|
|
chr12_+_22778085
|
0.621
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chr4_+_6675798
|
0.617
|
|
LOC93622
|
Morf4 family associated protein 1-like 1 pseudogene
|
|
chr11_+_9685624
|
0.613
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr12_+_104850762
|
0.613
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr16_+_57653649
|
0.610
|
|
GPR56
|
G protein-coupled receptor 56
|
|
chr15_-_55700456
|
0.609
|
NM_004748
NM_001204450
NM_001204451
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chr1_+_27153130
|
0.604
|
NM_032283
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr4_-_10118500
|
0.600
|
NM_005112
NM_017491
|
WDR1
|
WD repeat domain 1
|
|
chr2_+_47168312
|
0.598
|
NM_020458
|
TTC7A
|
tetratricopeptide repeat domain 7A
|
|
chr3_+_52529351
|
0.598
|
NM_015136
|
STAB1
|
stabilin 1
|
|
chr19_+_56652529
|
0.592
|
NM_001253792
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr1_+_151584514
|
0.584
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr13_+_52158592
|
0.584
|
|
WDFY2
|
WD repeat and FYVE domain containing 2
|
|
chr3_+_9944511
|
0.583
|
NM_001193380
NM_153480
|
IL17RE
|
interleukin 17 receptor E
|
|
chr19_-_2456954
|
0.582
|
NM_032737
|
LMNB2
|
lamin B2
|
|
chr19_-_4817502
|
0.579
|
|
TICAM1
|
toll-like receptor adaptor molecule 1
|
|
chrX_-_109561298
|
0.579
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr2_+_241544819
|
0.579
|
NM_001195381
NM_001195382
|
GPR35
|
G protein-coupled receptor 35
|
|
chr11_-_44972227
|
0.575
|
NM_001076787
NM_006034
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr16_-_14788429
|
0.572
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr7_+_48128738
|
0.571
|
NM_003364
|
UPP1
|
uridine phosphorylase 1
|
|
chr3_+_14166582
|
0.571
|
|
TMEM43
|
transmembrane protein 43
|
|
chrX_-_106959597
|
0.571
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr18_+_3594056
|
0.566
|
|
FLJ35776
|
uncharacterized LOC649446
|