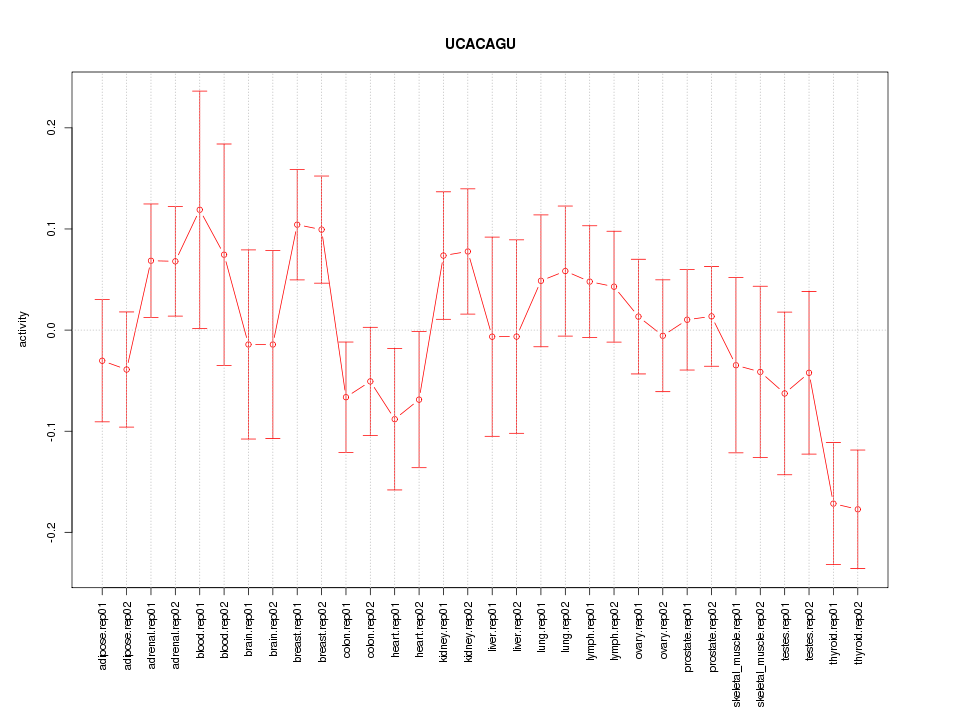
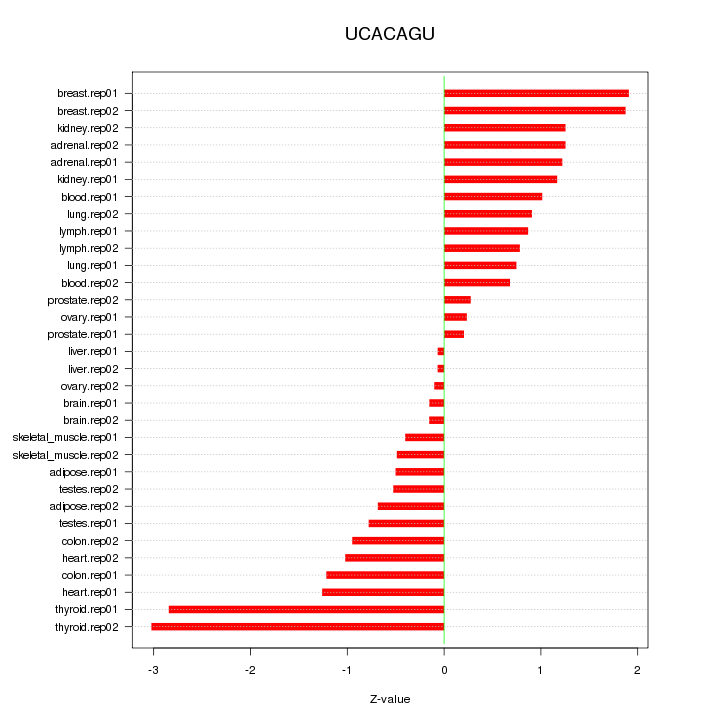
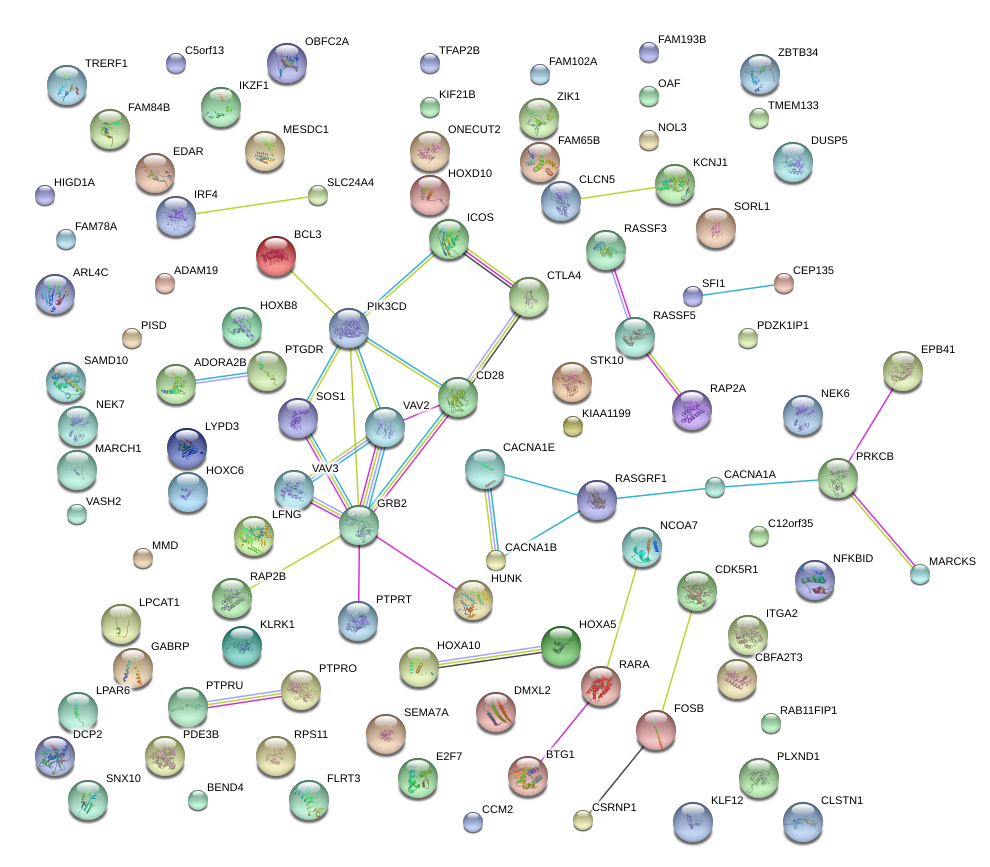
|
chr7_+_50344255
|
3.832
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr7_-_27183225
|
2.837
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr6_+_391738
|
2.747
|
NM_001195286
NM_002460
|
IRF4
|
interferon regulatory factor 4
|
|
chr19_+_45971249
|
2.591
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr2_+_204801453
|
2.563
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr6_+_50786438
|
2.295
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr7_-_27213873
|
2.244
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr2_-_235405692
|
2.212
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr1_-_108507474
|
2.088
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr6_-_24911194
|
2.088
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr1_-_200992824
|
2.086
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr11_+_121322848
|
2.062
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr1_-_47655723
|
2.050
|
NM_005764
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr2_+_204571197
|
2.012
|
NM_001243077
NM_001243078
NM_006139
|
CD28
|
CD28 molecule
|
|
chr2_-_109605668
|
1.990
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr1_-_108231122
|
1.966
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr14_+_92788924
|
1.916
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr14_+_92790151
|
1.890
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr9_-_130712866
|
1.889
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr14_+_92789536
|
1.847
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr3_+_152880024
|
1.822
|
NM_002886
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr17_+_30813928
|
1.741
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr19_-_36391448
|
1.702
|
NM_139239
|
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta
|
|
chr19_+_45251933
|
1.679
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr15_-_74726258
|
1.669
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr5_+_170210711
|
1.656
|
NM_014211
|
GABRP
|
gamma-aminobutyric acid (GABA) A receptor, pi
|
|
chr11_+_14665268
|
1.636
|
NM_000922
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr6_+_126111782
|
1.612
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr5_-_1524022
|
1.610
|
NM_024830
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr2_+_192542860
|
1.568
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr15_+_81071708
|
1.552
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr5_+_112312392
|
1.547
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr15_-_74726000
|
1.533
|
NM_001146030
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr4_-_164534656
|
1.499
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr17_-_46692300
|
1.493
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr3_-_129325175
|
1.474
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr7_+_26403944
|
1.468
|
NM_001199838
|
SNX10
|
sorting nexin 10
|
|
chr7_+_26331493
|
1.459
|
NM_001199835
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr1_+_206680863
|
1.456
|
NM_182663
NM_182664
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr9_-_130742803
|
1.433
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr17_-_53499055
|
1.426
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr5_-_111091947
|
1.423
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr17_-_73401788
|
1.403
|
NM_002086
NM_203506
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr9_+_129622935
|
1.378
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr15_+_81293286
|
1.375
|
NM_022566
|
MESDC1
|
mesoderm development candidate 1
|
|
chr4_-_42154894
|
1.373
|
NM_001159547
NM_207406
|
BEND4
|
BEN domain containing 4
|
|
chr7_+_26400503
|
1.371
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr13_-_48987652
|
1.358
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr1_+_29213569
|
1.328
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr14_+_52734430
|
1.325
|
NM_000953
|
PTGDR
|
prostaglandin D2 receptor (DP)
|
|
chr12_-_92539614
|
1.319
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr9_+_127020462
|
1.310
|
NM_001166168
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr12_+_54422193
|
1.291
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr10_+_112257624
|
1.251
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr6_+_126102278
|
1.251
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr6_+_114178516
|
1.250
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr9_+_127019782
|
1.223
|
NM_001166167
NM_001145001
NM_014397
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr1_+_9711781
|
1.211
|
NM_005026
|
PIK3CD
|
phosphoinositide-3-kinase, catalytic, delta polypeptide
|
|
chr5_-_171615320
|
1.199
|
NM_005990
|
STK10
|
serine/threonine kinase 10
|
|
chr11_+_120081669
|
1.192
|
NM_178507
|
OAF
|
OAF homolog (Drosophila)
|
|
chr4_-_165304406
|
1.181
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr16_-_89007605
|
1.159
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr8_-_127570710
|
1.149
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr19_-_43969723
|
1.148
|
NM_014400
|
LYPD3
|
LY6/PLAUR domain containing 3
|
|
chr7_+_2557496
|
1.148
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr9_+_127054567
|
1.143
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr20_-_14318200
|
1.129
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr12_+_65004191
|
1.125
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr13_-_74707913
|
1.120
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr20_-_41818464
|
1.114
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chrX_+_49832214
|
1.111
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr20_-_62610990
|
1.106
|
NM_080621
|
SAMD10
|
sterile alpha motif domain containing 10
|
|
chr17_+_15848230
|
1.106
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chr6_+_126240369
|
1.103
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr12_+_32112286
|
1.094
|
NM_018169
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr5_-_157002739
|
1.090
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr18_+_55102777
|
1.081
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr5_+_52284903
|
1.081
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr5_-_176981537
|
1.071
|
NM_001190946
|
FAM193B
|
family with sequence similarity 193, member B
|
|
chr21_+_33245304
|
1.070
|
NM_014586
|
HUNK
|
hormonally up-regulated Neu-associated kinase
|
|
chr11_-_128712340
|
1.058
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr4_+_56815036
|
1.042
|
NM_025009
|
CEP135
|
centrosomal protein 135kDa
|
|
chr7_+_2552162
|
1.032
|
NM_001166355
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr9_+_127054246
|
1.028
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr8_-_37756971
|
1.028
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr13_-_49018802
|
1.024
|
NM_005767
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr15_-_79383080
|
1.022
|
NM_001145648
NM_002891
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr16_+_23847271
|
1.018
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr22_-_32026783
|
1.017
|
NM_014338
|
PISD
|
phosphatidylserine decarboxylase
|
|
chr12_+_15699277
|
1.011
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr2_-_39347562
|
1.004
|
NM_005633
|
SOS1
|
son of sevenless homolog 1 (Drosophila)
|
|
chr3_-_39195101
|
1.002
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr19_-_13617037
|
0.998
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr15_-_51914770
|
0.975
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chr1_+_213123853
|
0.974
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr17_+_38474472
|
0.968
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr9_-_134151905
|
0.964
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr9_-_136857285
|
0.956
|
NM_001134398
NM_003371
|
VAV2
|
vav 2 guanine nucleotide exchange factor
|
|
chr13_-_49001042
|
0.955
|
NM_001162497
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr6_-_42419782
|
0.951
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr12_-_77459198
|
0.950
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr7_+_45039777
|
0.946
|
NM_001167934
NM_001167935
NM_031443
|
CCM2
|
cerebral cavernous malformation 2
|
|
chr6_+_11538486
|
0.933
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr7_+_7222142
|
0.931
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr6_+_41606193
|
0.929
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chrX_+_17393537
|
0.920
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr1_+_28585959
|
0.918
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr10_+_8096633
|
0.909
|
NM_001002295
NM_002051
|
GATA3
|
GATA binding protein 3
|
|
chr14_+_103243813
|
0.902
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr8_-_60031545
|
0.898
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr4_+_26862312
|
0.889
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr2_-_201936389
|
0.888
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr20_+_3776385
|
0.880
|
NM_004358
NM_021872
NM_021873
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr3_-_32023237
|
0.851
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr8_-_103136486
|
0.849
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr21_-_43373938
|
0.846
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr14_-_78083109
|
0.845
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr11_-_128737132
|
0.841
|
NM_153764
NM_153765
NM_153766
NM_153767
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr12_-_25403824
|
0.837
|
NM_004985
NM_033360
|
KRAS
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
|
|
chr15_-_79298001
|
0.836
|
NM_153815
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr10_+_11047253
|
0.834
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr12_+_53773924
|
0.833
|
NM_001251825
NM_138473
|
SP1
|
Sp1 transcription factor
|
|
chr5_-_83016850
|
0.833
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr17_-_58603492
|
0.832
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr8_-_22550708
|
0.831
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr12_+_99038918
|
0.829
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr16_-_70472920
|
0.828
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr1_+_110168048
|
0.823
|
NM_203404
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr17_-_56084656
|
0.819
|
NM_001078166
NM_006924
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr12_+_53774422
|
0.818
|
NM_003109
|
SP1
|
Sp1 transcription factor
|
|
chr16_-_89043215
|
0.816
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr12_-_106641675
|
0.814
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr1_+_228194722
|
0.812
|
NM_033131
|
WNT3A
|
wingless-type MMTV integration site family, member 3A
|
|
chr4_+_38665587
|
0.802
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr12_-_22487566
|
0.795
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr3_-_128206704
|
0.790
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr1_+_101361589
|
0.790
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr8_+_80523048
|
0.788
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr2_+_163200582
|
0.779
|
NM_012198
|
GCA
|
grancalcin, EF-hand calcium binding protein
|
|
chr8_-_22550057
|
0.774
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr1_+_244216506
|
0.773
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr4_+_77870864
|
0.769
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr1_+_199996729
|
0.766
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr11_+_122526339
|
0.766
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr7_-_139876718
|
0.764
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr2_+_113403433
|
0.753
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr16_+_577833
|
0.753
|
NM_005632
|
SOLH
|
small optic lobes homolog (Drosophila)
|
|
chr17_-_28562953
|
0.752
|
NM_001045
|
SLC6A4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4
|
|
chr19_+_11200037
|
0.752
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr2_+_85132762
|
0.750
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr1_-_161039745
|
0.749
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr19_-_47734215
|
0.744
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr12_-_120703549
|
0.741
|
NM_001080855
NM_001243756
NM_002859
|
PXN
|
paxillin
|
|
chr11_-_102826433
|
0.739
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chrX_+_49687212
|
0.737
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr10_-_60027630
|
0.736
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr3_-_128212027
|
0.731
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr10_+_11206928
|
0.726
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr11_-_88796815
|
0.724
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr1_-_93646245
|
0.720
|
NM_001167830
NM_016040
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr16_-_48644087
|
0.719
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr17_+_25621105
|
0.719
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr17_-_40075238
|
0.718
|
NM_001096
NM_198830
|
ACLY
|
ATP citrate lyase
|
|
chr12_+_105501448
|
0.717
|
NM_015275
|
KIAA1033
|
KIAA1033
|
|
chr6_-_149806116
|
0.717
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr1_-_39338976
|
0.717
|
NM_012333
|
MYCBP
|
c-myc binding protein
|
|
chr8_+_125486949
|
0.715
|
NM_007218
|
RNF139
|
ring finger protein 139
|
|
chr16_-_11680146
|
0.712
|
NM_004862
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr17_+_37026111
|
0.712
|
NM_006148
|
LASP1
|
LIM and SH3 protein 1
|
|
chr18_-_74207145
|
0.711
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr5_+_14664764
|
0.709
|
NM_138348
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr16_-_68482373
|
0.707
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr18_-_3011944
|
0.704
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr2_+_74273449
|
0.703
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr12_-_65153189
|
0.697
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr1_+_23037329
|
0.695
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr1_+_206730456
|
0.693
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr14_+_56584854
|
0.692
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr13_+_24734788
|
0.690
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr8_-_22549901
|
0.688
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_74791098
|
0.688
|
NM_001001481
NM_018299
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr7_-_92465929
|
0.687
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chrX_-_110654373
|
0.684
|
NM_000555
|
DCX
|
doublecortin
|
|
chr14_-_39901619
|
0.683
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr7_+_30174343
|
0.682
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr4_+_174089889
|
0.681
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr20_-_20693122
|
0.675
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr7_+_6144504
|
0.675
|
NM_032172
|
USP42
|
ubiquitin specific peptidase 42
|
|
chr16_-_11680756
|
0.672
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr12_+_54410641
|
0.671
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr12_+_15475190
|
0.656
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr5_+_82767492
|
0.655
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr8_-_103137134
|
0.650
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr1_+_207070787
|
0.649
|
NM_001185156
NM_001185157
NM_001185158
NM_006850
|
IL24
|
interleukin 24
|
|
chr17_+_38498270
|
0.648
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr18_+_13218777
|
0.644
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr17_+_38278789
|
0.643
|
NM_001012241
|
MSL1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr9_+_130830447
|
0.643
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr18_+_13620866
|
0.643
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr9_+_137533643
|
0.641
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|