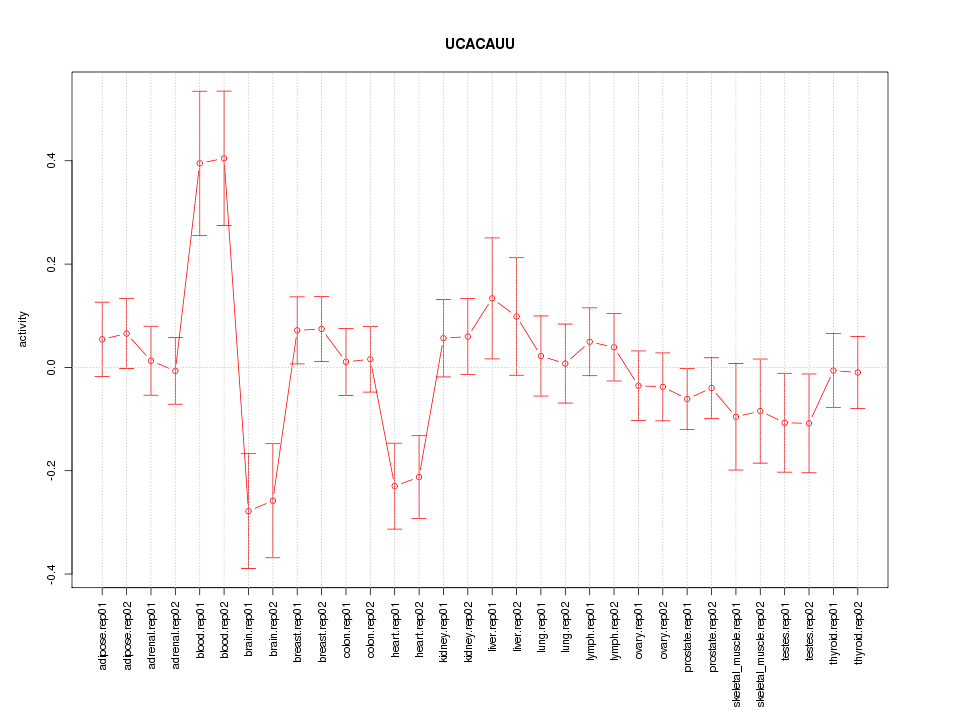
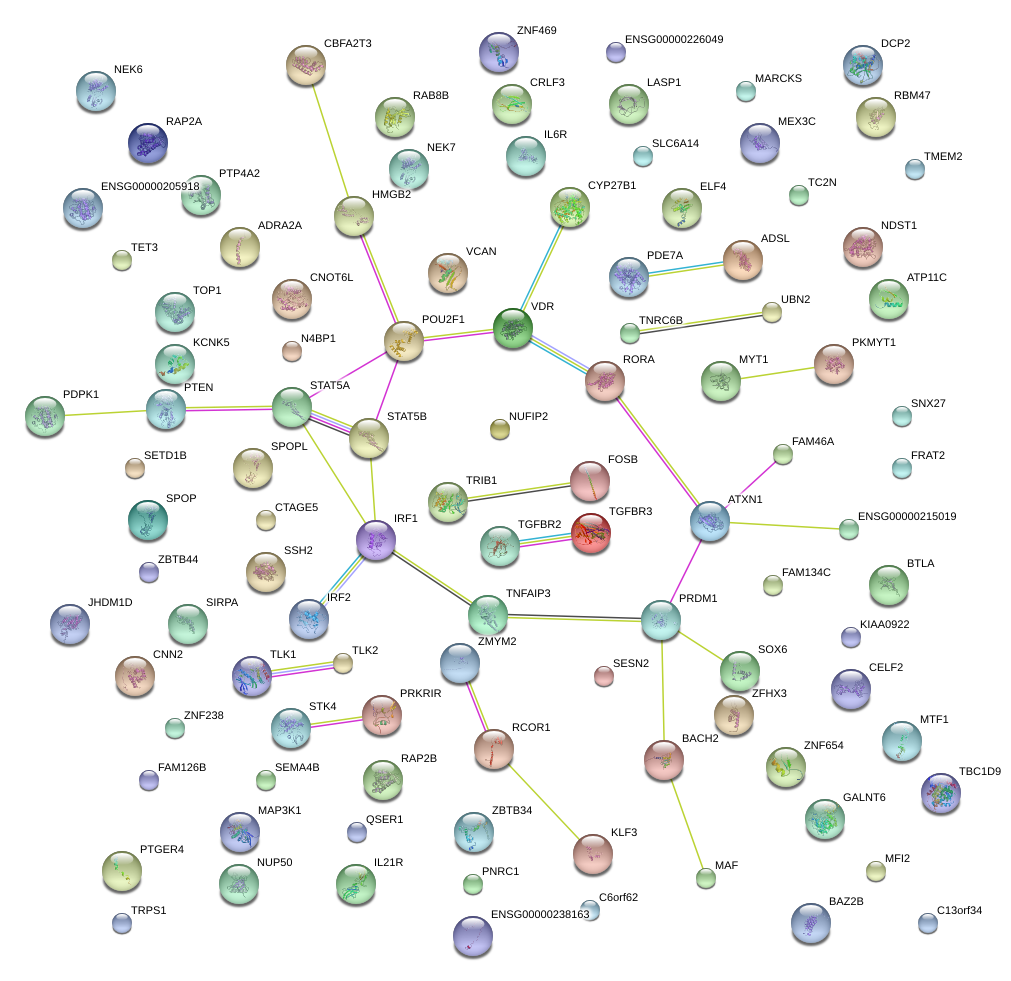
|
chr4_-_40631846
|
6.437
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr4_-_40517913
|
4.842
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr5_+_40680021
|
4.655
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr17_-_29151724
|
4.352
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr19_+_1026271
|
4.337
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr22_+_40573879
|
4.312
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr7_-_139876718
|
4.209
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr4_-_78740508
|
4.146
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr9_-_74383301
|
4.136
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr9_+_127020462
|
4.043
|
NM_001166168
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr9_+_127019782
|
4.012
|
NM_001166167
NM_001145001
NM_014397
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr4_+_38665587
|
4.001
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr3_+_30647901
|
3.794
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr16_+_27413482
|
3.658
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr16_-_89007605
|
3.653
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr5_+_56110899
|
3.534
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr9_+_127054567
|
3.529
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr5_+_82767492
|
3.438
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr15_-_60884615
|
3.433
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chrX_-_129244652
|
3.290
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr6_-_39197225
|
3.260
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr5_-_131826426
|
3.253
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr9_+_127054246
|
3.252
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr22_+_40440664
|
3.233
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr9_+_129622935
|
3.232
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr15_-_61521478
|
3.216
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chrX_-_129244471
|
3.118
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr17_-_40428391
|
3.060
|
NM_012448
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr6_+_114178516
|
2.991
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr10_+_11047253
|
2.979
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr22_+_45560529
|
2.974
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr22_+_45559725
|
2.946
|
NM_007172
|
NUP50
|
nucleoporin 50kDa
|
|
chr16_-_89043215
|
2.941
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr6_+_89790412
|
2.896
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr19_+_45971249
|
2.864
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr2_+_74273449
|
2.863
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr6_-_91006460
|
2.791
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr6_-_91006580
|
2.776
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr10_+_11206928
|
2.761
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr8_+_126442557
|
2.720
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr16_+_27438578
|
2.703
|
NM_021798
|
IL21R
|
interleukin 21 receptor
|
|
chr1_+_244216506
|
2.630
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr20_+_1875825
|
2.507
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr4_+_154387450
|
2.486
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr15_+_63481713
|
2.473
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr16_-_48644087
|
2.472
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr12_-_48298765
|
2.468
|
NM_000376
NM_001017535
NM_001017536
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr20_+_43595119
|
2.418
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr17_-_27621163
|
2.415
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr12_+_122242580
|
2.349
|
NM_015048
|
SETD1B
|
SET domain containing 1B
|
|
chr20_+_1875382
|
2.307
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr1_+_167298280
|
2.289
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr14_+_103058988
|
2.263
|
NM_015156
|
RCOR1
|
REST corepressor 1
|
|
chr18_-_48724031
|
2.213
|
NM_016626
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr11_+_32914359
|
2.189
|
NM_001076786
|
QSER1
|
glutamine and serine rich 1
|
|
chr15_-_60919642
|
2.166
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr8_-_66701175
|
2.106
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr20_+_39657461
|
2.083
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr1_-_92351613
|
2.058
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr13_+_73301980
|
2.049
|
NM_024808
|
BORA
|
bora, aurora kinase A activator
|
|
chr10_+_11059854
|
2.042
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_32385258
|
2.006
|
NM_001195100
NM_001195101
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr3_+_88188261
|
1.999
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr1_+_154377668
|
1.984
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr6_-_16761600
|
1.970
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr6_-_24719286
|
1.963
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr5_+_149887673
|
1.962
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr2_-_171923482
|
1.962
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr14_+_39734475
|
1.915
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr14_+_39735500
|
1.907
|
NM_001247988
NM_203356
|
CTAGE5
|
CTAGE family, member 5
|
|
chr6_+_138188352
|
1.900
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr14_+_39736277
|
1.899
|
NM_001247989
NM_001247990
NM_005930
NM_203355
|
CTAGE5
|
CTAGE family, member 5
|
|
chr11_-_76091866
|
1.889
|
NM_004705
|
PRKRIR
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor)
|
|
chr4_-_185395616
|
1.871
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr8_-_66753968
|
1.842
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr6_+_106546736
|
1.841
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr7_+_138916230
|
1.832
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr16_-_73092533
|
1.807
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr3_+_152880024
|
1.806
|
NM_002886
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr16_+_2587951
|
1.755
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr5_+_112312392
|
1.749
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr6_+_106534188
|
1.742
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr8_-_116681103
|
1.739
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr17_-_40761404
|
1.728
|
NM_178126
|
FAM134C
|
family with sequence similarity 134, member C
|
|
chrX_+_115567746
|
1.721
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr2_-_201936389
|
1.721
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr15_+_90728118
|
1.687
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr4_-_174254730
|
1.683
|
NM_001130689
|
HMGB2
|
high mobility group box 2
|
|
chr4_-_141677470
|
1.682
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr1_+_28585959
|
1.651
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr10_-_99094427
|
1.649
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr11_-_16424391
|
1.646
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr2_-_172087823
|
1.603
|
NM_001136554
|
TLK1
|
tousled-like kinase 1
|
|
chr11_-_130184459
|
1.603
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr10_+_89623091
|
1.598
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr1_+_244214552
|
1.596
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_167189948
|
1.596
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr2_-_160472960
|
1.592
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr19_-_33793429
|
1.590
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr17_+_37026111
|
1.551
|
NM_006148
|
LASP1
|
LIM and SH3 protein 1
|
|
chr16_-_79634610
|
1.543
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr3_-_112218383
|
1.539
|
NM_001085357
NM_181780
|
BTLA
|
B and T lymphocyte associated
|
|
chr16_+_88493878
|
1.535
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr13_+_20532906
|
1.526
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chrX_-_138914384
|
1.490
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr17_-_28257017
|
1.437
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr10_+_112836784
|
1.423
|
NM_000681
|
ADRA2A
|
adrenergic, alpha-2A-, receptor
|
|
chr6_-_82462374
|
1.402
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr7_-_106301329
|
1.399
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr1_+_151584514
|
1.393
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr2_-_172017358
|
1.388
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr2_+_139259349
|
1.380
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr1_-_38325224
|
1.347
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr1_+_26798901
|
1.343
|
NM_005517
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr11_-_85376159
|
1.330
|
NM_001039618
|
CREBZF
|
CREB/ATF bZIP transcription factor
|
|
chr1_+_44115795
|
1.328
|
NM_014663
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr12_+_22778008
|
1.320
|
NM_001039481
NM_018638
|
ETNK1
|
ethanolamine kinase 1
|
|
chr2_+_112812799
|
1.317
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr2_-_206950895
|
1.316
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr5_-_77944341
|
1.312
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr3_+_114012832
|
1.311
|
NM_173799
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains
|
|
chr1_+_66458389
|
1.284
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr20_+_1874779
|
1.281
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr2_-_128100673
|
1.278
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr20_-_31071187
|
1.266
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr2_-_225450105
|
1.259
|
NM_003590
|
CUL3
|
cullin 3
|
|
chr15_+_100173182
|
1.256
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chrX_-_124097601
|
1.244
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr7_+_114562140
|
1.243
|
NM_001166345
NM_001166346
NM_199072
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr12_-_48213567
|
1.233
|
NM_001098416
NM_015401
|
HDAC7
|
histone deacetylase 7
|
|
chr18_+_60190657
|
1.230
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr12_-_80328926
|
1.219
|
NM_002480
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr2_+_152214105
|
1.215
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr4_-_140060601
|
1.211
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr16_-_73082169
|
1.204
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr6_-_64029847
|
1.197
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr9_+_101569933
|
1.194
|
NM_024642
|
GALNT12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12)
|
|
chr3_-_157213138
|
1.194
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chrX_+_44732402
|
1.175
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr1_+_236558715
|
1.165
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr1_-_212004113
|
1.160
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr11_+_124609775
|
1.155
|
NM_001126181
NM_006176
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr17_-_47439326
|
1.151
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr13_+_20532788
|
1.149
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr1_-_32403975
|
1.129
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr5_+_72112417
|
1.123
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chrX_-_63425484
|
1.115
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr1_+_63833260
|
1.114
|
NM_013339
|
ALG6
|
asparagine-linked glycosylation 6, alpha-1,3-glucosyltransferase homolog (S. cerevisiae)
|
|
chr6_+_119215240
|
1.111
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr3_+_196466724
|
1.109
|
NM_002577
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr17_+_76000248
|
1.102
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr2_+_135676387
|
1.101
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr10_+_99079017
|
1.095
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr7_+_98476084
|
1.090
|
NM_001244580
NM_003496
|
TRRAP
|
transformation/transcription domain-associated protein
|
|
chr8_-_28243941
|
1.069
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr17_-_66287256
|
1.068
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr4_+_41937105
|
1.064
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chr1_+_66258855
|
1.060
|
NM_002600
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr10_-_60027630
|
1.060
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr16_-_17564737
|
1.032
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr17_+_75181294
|
1.028
|
NM_001144001
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr12_+_104680712
|
1.025
|
NM_003330
NM_182729
NM_182742
NM_182743
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr1_-_240775448
|
1.020
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr10_-_101492422
|
1.019
|
NM_004376
NM_078470
|
COX15
|
COX15 homolog, cytochrome c oxidase assembly protein (yeast)
|
|
chr12_+_104850641
|
1.017
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr3_+_33155447
|
1.003
|
NM_006371
|
CRTAP
|
cartilage associated protein
|
|
chr16_+_24740945
|
1.002
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr1_+_66258192
|
1.001
|
NM_001037341
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr17_+_75136985
|
0.993
|
NM_001039573
NM_001143998
NM_001143999
NM_003003
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr11_+_94277005
|
0.992
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr12_-_80329234
|
0.978
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr1_-_92371558
|
0.954
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr16_+_53468344
|
0.951
|
NM_005611
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr5_-_78282356
|
0.921
|
NM_000046
|
ARSB
|
arylsulfatase B
|
|
chr3_+_63884074
|
0.910
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr11_-_47510518
|
0.910
|
NM_001025596
NM_001172640
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr17_-_65241305
|
0.900
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr6_-_8064642
|
0.889
|
NM_001199322
NM_001199323
NM_201280
|
MUTED
|
muted homolog (mouse)
|
|
chr1_-_93646245
|
0.887
|
NM_001167830
NM_016040
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr5_+_72143929
|
0.887
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr1_-_225615787
|
0.879
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chr1_-_154155698
|
0.868
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr7_-_115670828
|
0.855
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr7_-_20256942
|
0.855
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr7_+_4721722
|
0.853
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr12_-_80307690
|
0.849
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr12_-_42538432
|
0.849
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr17_+_28705922
|
0.846
|
NM_001304
|
CPD
|
carboxypeptidase D
|
|
chr17_+_75085234
|
0.845
|
NM_001204410
|
SEC14L1
LINC00338
|
SEC14-like 1 (S. cerevisiae)
long intergenic non-protein coding RNA 338
|
|
chr4_-_174255454
|
0.837
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chr12_+_122150638
|
0.836
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr6_+_10521564
|
0.834
|
NM_145649
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr21_+_44394634
|
0.829
|
NM_004571
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr17_+_1959512
|
0.823
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr22_-_32146119
|
0.823
|
NM_173566
|
PRR14L
|
proline rich 14-like
|
|
chr17_+_61699786
|
0.816
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr11_+_67033889
|
0.801
|
NM_001619
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr12_-_10766130
|
0.799
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr2_-_47143250
|
0.795
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr17_-_43502992
|
0.795
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|