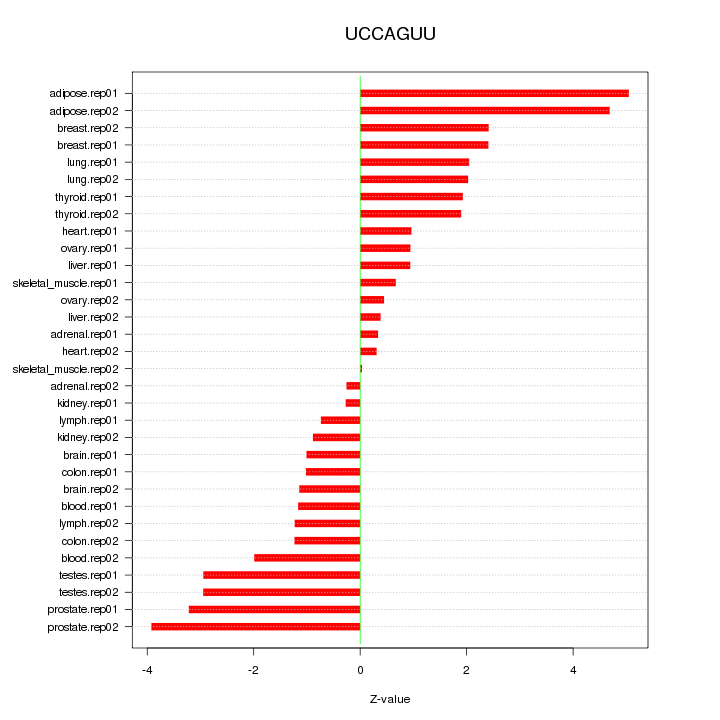
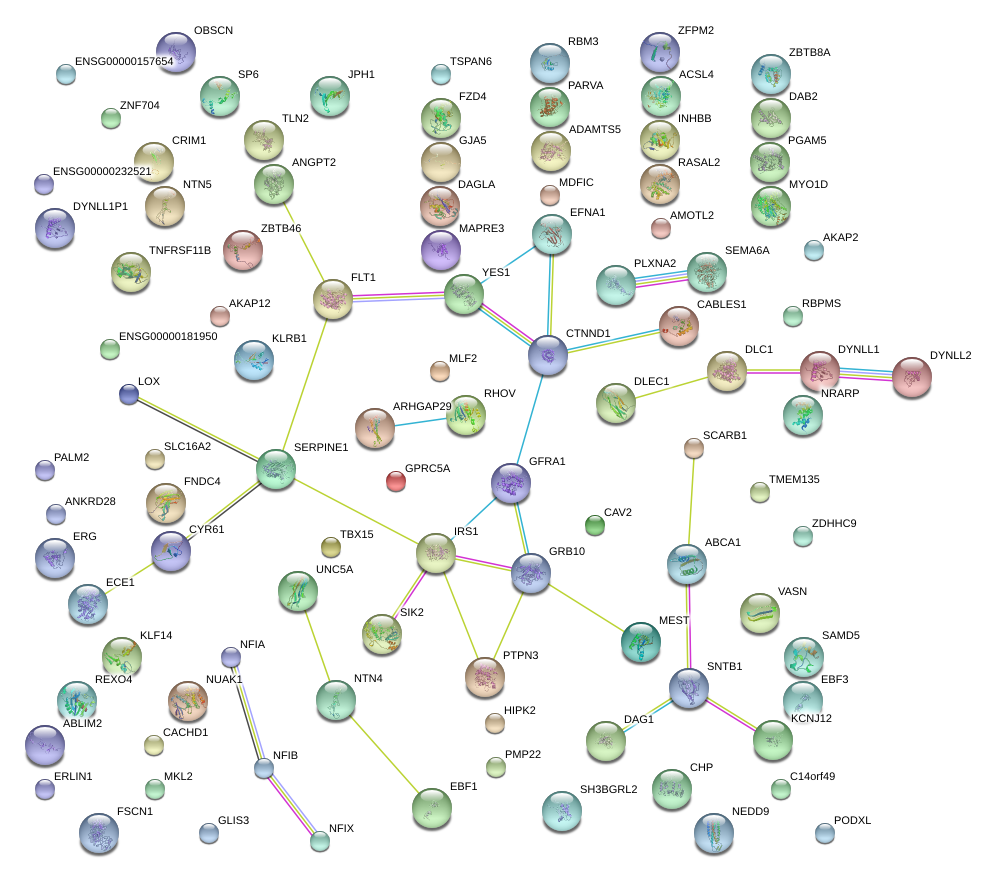
|
chr2_+_121103670
|
4.457
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr1_+_86046434
|
4.429
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr21_-_39870303
|
4.191
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr7_+_116139619
|
3.986
|
NM_001206747
NM_001233
NM_198212
|
CAV2
|
caveolin 2
|
|
chr8_-_12990731
|
3.682
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr8_-_12973752
|
3.645
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr11_-_86666211
|
3.555
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr8_-_119964059
|
3.540
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr7_+_116140097
|
3.455
|
NM_001206748
|
CAV2
|
caveolin 2
|
|
chr7_-_131241321
|
3.390
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr9_+_112403067
|
3.316
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr5_-_121412805
|
3.288
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr9_-_112213663
|
3.034
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr9_-_112191105
|
3.023
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chrX_-_99891690
|
3.022
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr1_-_147232684
|
3.007
|
NM_181703
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr1_-_147245483
|
2.983
|
NM_005266
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr6_+_151646634
|
2.945
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr9_-_14398981
|
2.901
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr21_-_28339405
|
2.818
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr21_-_40033617
|
2.815
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_-_94703252
|
2.678
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr5_-_121414011
|
2.669
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr8_-_6420454
|
2.629
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr10_-_118031542
|
2.575
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr9_-_14314036
|
2.523
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr16_+_4421774
|
2.496
|
NM_138440
|
VASN
|
vasorin
|
|
chr5_-_115910406
|
2.494
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr7_+_130131921
|
2.485
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr12_-_106533810
|
2.474
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr10_-_118032717
|
2.411
|
NM_001145453
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr4_-_8160435
|
2.391
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr8_-_81787015
|
2.369
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr9_-_112260529
|
2.346
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr11_+_12399024
|
2.238
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr7_+_130126035
|
2.217
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr10_-_118032975
|
2.211
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr3_-_134093235
|
2.202
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr7_-_139477437
|
2.172
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chrX_+_73641084
|
2.153
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr12_-_96184510
|
2.124
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr12_-_125348422
|
2.114
|
NM_001082959
NM_005505
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr17_-_15165873
|
2.085
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr17_-_15168590
|
2.078
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr7_+_130131172
|
2.075
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr9_-_107690526
|
2.074
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr8_-_13372183
|
2.070
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr18_-_812268
|
2.027
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr20_-_62436855
|
1.995
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr10_-_131762017
|
1.978
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr2_-_227663454
|
1.970
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr6_+_151561094
|
1.951
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_-_11382501
|
1.895
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr1_+_61542928
|
1.869
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr11_+_111473099
|
1.864
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr7_+_5632435
|
1.852
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr17_-_15164043
|
1.832
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr1_+_61547388
|
1.787
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_155100304
|
1.719
|
NM_004428
NM_182685
|
EFNA1
|
ephrin-A1
|
|
chr5_-_39425297
|
1.705
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr1_-_119532178
|
1.687
|
NM_152380
|
TBX15
|
T-box 15
|
|
chr11_+_86748870
|
1.685
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr7_+_100770378
|
1.669
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr13_-_29069215
|
1.625
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr10_-_101945787
|
1.588
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chrX_+_48432740
|
1.550
|
NM_006743
|
RBM3
|
RNA binding motif (RNP1, RRM) protein 3
|
|
chr1_+_64936475
|
1.543
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr15_+_62939509
|
1.535
|
NM_015059
|
TLN2
|
talin 2
|
|
chr6_+_147829784
|
1.534
|
NM_001030060
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr3_-_15798057
|
1.503
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr2_-_27718098
|
1.499
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr8_+_30241943
|
1.499
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr1_-_21671905
|
1.496
|
NM_001113348
|
ECE1
|
endothelin converting enzyme 1
|
|
chr10_-_101945686
|
1.483
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr1_-_21616648
|
1.447
|
NM_001113349
|
ECE1
|
endothelin converting enzyme 1
|
|
chr1_+_61547625
|
1.445
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr7_-_50800018
|
1.436
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr6_+_80340971
|
1.432
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr1_-_21605984
|
1.426
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr1_-_21616888
|
1.419
|
NM_001397
|
ECE1
|
endothelin converting enzyme 1
|
|
chr5_-_158526698
|
1.413
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr8_+_106331146
|
1.377
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr11_+_57529233
|
1.362
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chr1_+_178062829
|
1.350
|
NM_170692
|
RASAL2
|
RAS protein activator like 2
|
|
chr7_+_114562140
|
1.350
|
NM_001166345
NM_001166346
NM_199072
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr15_+_41523401
|
1.349
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr8_-_121824287
|
1.346
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr7_-_50772997
|
1.338
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr16_+_14164879
|
1.322
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr9_+_112542496
|
1.319
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr1_-_208417635
|
1.309
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr2_+_36583369
|
1.309
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr18_+_20735788
|
1.306
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr14_-_95942037
|
1.295
|
NM_152592
|
C14orf49
|
chromosome 14 open reading frame 49
|
|
chr17_-_31203889
|
1.279
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chrX_-_108976509
|
1.256
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr18_+_20715726
|
1.230
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr3_+_49506079
|
1.216
|
NM_001177643
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr7_-_130418859
|
1.207
|
NM_138693
|
KLF14
|
Kruppel-like factor 14
|
|
chr1_+_33004771
|
1.203
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr9_-_140196702
|
1.196
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chrX_-_128977719
|
1.191
|
NM_016032
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr8_-_75233461
|
1.182
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr12_+_13043955
|
1.176
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr11_+_61447816
|
1.175
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chrX_-_128977291
|
1.175
|
NM_001008222
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr9_-_4300028
|
1.173
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr17_+_21279667
|
1.169
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr1_+_78956727
|
1.167
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|
|
chr19_-_11308184
|
1.164
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr9_-_4152182
|
1.160
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr12_+_64238511
|
1.157
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr7_-_79082870
|
1.124
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr3_+_171757413
|
1.114
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr18_-_22932109
|
1.113
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr19_-_11305186
|
1.092
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr3_+_57994087
|
1.090
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr17_+_46125675
|
1.083
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr16_-_49860917
|
1.071
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr21_+_40177847
|
1.071
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr19_+_34745452
|
1.063
|
NM_014686
|
KIAA0355
|
KIAA0355
|
|
chr2_-_242255114
|
1.062
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr12_+_10365415
|
1.046
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr14_-_23623493
|
1.039
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr11_-_128457452
|
1.037
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr19_-_16683188
|
1.032
|
NM_024881
|
SLC35E1
|
solute carrier family 35, member E1
|
|
chr3_-_195808997
|
1.029
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr1_+_178310605
|
1.025
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr12_-_89746244
|
1.019
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr4_+_160188997
|
1.018
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr12_+_131438451
|
1.012
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr6_-_139695784
|
0.996
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr12_-_120703549
|
0.996
|
NM_001080855
NM_001243756
NM_002859
|
PXN
|
paxillin
|
|
chr6_+_41606193
|
0.993
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr3_-_195808958
|
0.990
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr18_+_55862577
|
0.987
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr16_+_19125243
|
0.984
|
NM_001034841
|
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2
|
|
chr14_-_23652840
|
0.973
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr6_-_139695349
|
0.966
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr13_+_113656027
|
0.943
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr3_-_129325175
|
0.936
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr17_+_7608413
|
0.925
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr1_+_76540374
|
0.924
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr3_+_121312169
|
0.923
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr13_-_41240607
|
0.916
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr6_-_111804413
|
0.913
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr3_+_111451326
|
0.903
|
NM_001134437
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr1_-_94374986
|
0.897
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr6_+_143929316
|
0.896
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr10_+_95848811
|
0.887
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr12_-_120687956
|
0.880
|
NM_025157
|
PXN
|
paxillin
|
|
chr6_-_11232901
|
0.874
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr3_+_111578026
|
0.872
|
NM_001134438
NM_145753
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr16_+_69220940
|
0.872
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr13_+_113623496
|
0.869
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr18_+_55714457
|
0.867
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr13_+_24144402
|
0.866
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr2_+_39893021
|
0.863
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr1_+_200708604
|
0.860
|
NM_203459
|
CAMSAP2
|
calmodulin regulated spectrin-associated protein family, member 2
|
|
chr14_-_35182944
|
0.853
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr12_+_58087737
|
0.845
|
NM_001017956
NM_001017957
NM_001017958
NM_006812
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin
|
|
chr7_-_2354050
|
0.844
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr4_-_114682836
|
0.840
|
NM_001221
NM_172114
NM_172115
NM_172127
NM_172128
NM_172129
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr1_+_160085519
|
0.836
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr3_+_49507470
|
0.836
|
NM_001165928
NM_001177634
NM_001177635
NM_001177636
NM_001177637
NM_001177638
NM_001177639
NM_001177640
NM_001177641
NM_001177642
NM_001177644
NM_004393
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr2_+_70485204
|
0.829
|
NM_016297
|
PCYOX1
|
prenylcysteine oxidase 1
|
|
chrX_+_119495939
|
0.829
|
NM_001142447
NM_012069
|
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide
|
|
chr15_-_65067696
|
0.828
|
NM_194272
|
RBPMS2
|
RNA binding protein with multiple splicing 2
|
|
chr20_+_51801821
|
0.823
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr14_-_61115906
|
0.819
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr15_+_67418053
|
0.808
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr8_-_23712297
|
0.805
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr7_-_83824216
|
0.803
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr6_+_7107799
|
0.801
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr18_+_3247527
|
0.798
|
NM_006471
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr17_-_40307015
|
0.795
|
NM_001252039
NM_004583
NM_201434
|
RAB5C
|
RAB5C, member RAS oncogene family
|
|
chr12_+_72233486
|
0.793
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr5_-_178772430
|
0.790
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr17_+_72983723
|
0.788
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr12_+_93771607
|
0.780
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr7_-_50861114
|
0.776
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr7_+_872137
|
0.770
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr5_-_179233772
|
0.768
|
NM_014275
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr12_+_50794591
|
0.766
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr3_+_111578528
|
0.762
|
NM_001134439
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr9_-_27529434
|
0.760
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr11_-_10590078
|
0.745
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr3_+_30647901
|
0.745
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr20_-_25566137
|
0.740
|
NM_025176
|
NINL
|
ninein-like
|
|
chr5_-_179229874
|
0.734
|
NM_054013
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr9_+_100263461
|
0.734
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr14_-_73493744
|
0.731
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr3_-_112359962
|
0.730
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr2_-_204399906
|
0.729
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr18_-_53255734
|
0.723
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chrX_-_10544852
|
0.721
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr8_-_27850368
|
0.718
|
NM_173833
|
SCARA5
|
scavenger receptor class A, member 5 (putative)
|
|
chr22_-_46933066
|
0.715
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr10_+_72163860
|
0.707
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr17_+_73452663
|
0.704
|
NM_014738
|
KIAA0195
|
KIAA0195
|