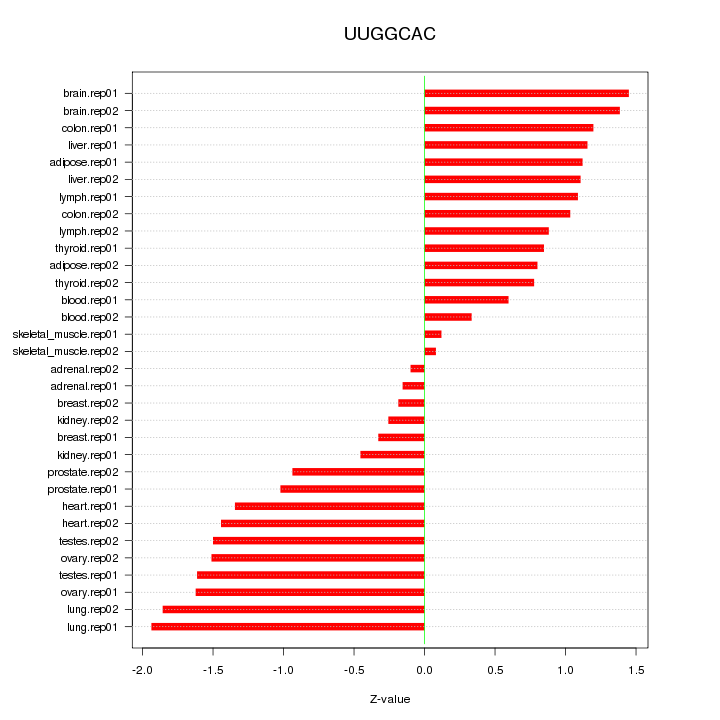
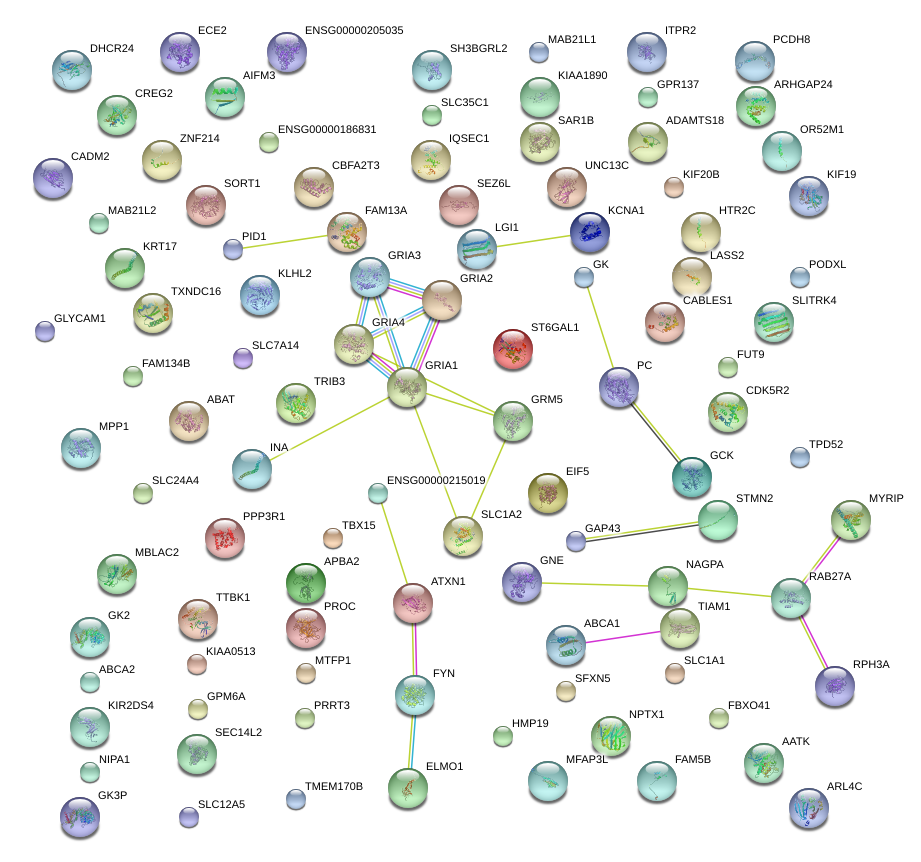
|
chr16_+_8814567
|
3.265
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr16_+_8806825
|
3.021
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr11_-_35441501
|
2.854
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr16_+_8768403
|
2.704
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr12_+_113229548
|
2.608
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr11_-_35441099
|
2.566
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr11_-_66675334
|
2.549
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr8_+_80523048
|
2.504
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr4_-_176923572
|
2.407
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr1_-_119532178
|
2.402
|
NM_152380
|
TBX15
|
T-box 15
|
|
chr5_+_173472692
|
2.370
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr22_+_26565424
|
2.283
|
NM_001184773
NM_001184774
NM_001184775
NM_001184776
NM_001184777
NM_021115
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr11_-_66725790
|
2.275
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr10_+_95517565
|
2.201
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr17_-_79105747
|
2.128
|
NM_004920
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr16_-_77468930
|
2.084
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr2_+_219824346
|
2.069
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr4_-_176734181
|
1.934
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr3_-_170303774
|
1.920
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (orphan transporter), member 14
|
|
chr19_-_33793429
|
1.883
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr3_+_186739643
|
1.876
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr17_-_79139871
|
1.874
|
NM_001080395
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr3_+_115342150
|
1.845
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr6_+_43211221
|
1.795
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr3_+_186648273
|
1.746
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr4_+_151503076
|
1.738
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr6_+_11538486
|
1.732
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr3_+_85775631
|
1.715
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr20_+_361272
|
1.695
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr6_+_80340971
|
1.690
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr22_+_21319417
|
1.681
|
NM_001018060
NM_144704
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr22_+_21321446
|
1.653
|
NM_001146288
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr20_+_44657812
|
1.650
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr9_+_4490193
|
1.568
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr8_-_81083660
|
1.565
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr20_+_44650260
|
1.565
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr5_+_152870083
|
1.536
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr15_-_55581969
|
1.485
|
NM_183235
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr2_-_73298815
|
1.482
|
NM_144579
|
SFXN5
|
sideroflexin 5
|
|
chr15_-_23086290
|
1.479
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr9_-_139923214
|
1.463
|
NM_212533
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr3_+_85008031
|
1.462
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr4_+_166131170
|
1.440
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr3_+_39851028
|
1.413
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr2_-_102003923
|
1.410
|
NM_153836
|
CREG2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr4_-_170924911
|
1.400
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr16_-_5083933
|
1.397
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr15_-_23086842
|
1.393
|
NM_001142275
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr7_-_37024664
|
1.392
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr13_-_53422773
|
1.379
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr11_+_45825622
|
1.346
|
NM_001145265
NM_001145266
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chr18_+_20715726
|
1.330
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr18_+_20735788
|
1.330
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr12_+_5019048
|
1.315
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr3_-_13114547
|
1.308
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr12_-_26985922
|
1.298
|
NM_002223
|
ITPR2
|
inositol 1,4,5-trisphosphate receptor, type 2
|
|
chrX_+_113818550
|
1.298
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr2_-_73496595
|
1.291
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr15_+_29213839
|
1.287
|
NM_001130414
NM_005503
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2
|
|
chr14_+_92788924
|
1.281
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr3_-_9994057
|
1.274
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr4_+_86851425
|
1.271
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_-_55352920
|
1.269
|
NM_014762
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr1_+_177140530
|
1.265
|
NM_021165
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr5_-_16509106
|
1.265
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr8_-_80993009
|
1.255
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr2_-_230135954
|
1.252
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr11_-_88796815
|
1.249
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr14_+_103801160
|
1.243
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr14_+_92790151
|
1.238
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr15_-_55562483
|
1.234
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr15_+_54305100
|
1.234
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr7_-_37393271
|
1.224
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr14_+_92789536
|
1.217
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr22_+_30792929
|
1.210
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr16_+_85061303
|
1.202
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr2_-_68479481
|
1.184
|
NM_000945
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr3_+_183993798
|
1.171
|
NM_001037324
NM_001100120
NM_001100121
|
ECE2
|
endothelin converting enzyme 2
|
|
chr10_+_105036783
|
1.164
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr1_-_109935869
|
1.160
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr5_-_89770432
|
1.153
|
NM_203406
|
MBLAC2
|
metallo-beta-lactamase domain containing 2
|
|
chr9_-_139922705
|
1.151
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr4_-_170947428
|
1.149
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr17_+_72322350
|
1.148
|
NM_153209
|
KIF19
|
kinesin family member 19
|
|
chr1_-_150947456
|
1.141
|
NM_022075
NM_181746
|
CERS2
|
ceramide synthase 2
|
|
chr6_+_96463844
|
1.140
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr4_-_89744154
|
1.133
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr9_-_107690526
|
1.131
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr2_-_235405692
|
1.128
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr4_+_86699846
|
1.125
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr6_-_112080284
|
1.110
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chrX_-_154033729
|
1.106
|
NM_001166460
NM_001166461
NM_001166462
NM_002436
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa
|
|
chr7_-_37488894
|
1.085
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr9_-_36258383
|
1.083
|
NM_001190383
NM_001190384
NM_005476
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr14_+_103800492
|
1.077
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr3_-_161089747
|
1.070
|
NM_001040100
|
SPTSSB
|
serine palmitoyltransferase, small subunit B
|
|
chr17_-_78450247
|
1.059
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chrX_-_142723921
|
1.045
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr6_-_16761600
|
1.041
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr16_-_89043215
|
1.035
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chrX_+_30671456
|
1.031
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chr6_-_112194648
|
1.030
|
NM_002037
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr6_-_112041253
|
1.025
|
NM_153047
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr21_-_32931289
|
1.022
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr5_-_133968463
|
1.021
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chr12_-_121734543
|
1.016
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr7_-_37488554
|
1.012
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr2_-_158731622
|
1.011
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr3_-_157823815
|
1.008
|
NM_001163678
NM_003030
NM_006884
|
SHOX2
|
short stature homeobox 2
|
|
chr8_+_67687415
|
1.006
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr6_+_42788793
|
1.000
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr1_-_211307314
|
0.998
|
NM_002238
NM_172362
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr18_+_13611664
|
0.987
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr2_+_166152282
|
0.982
|
NM_001040143
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr16_-_52581713
|
0.979
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr9_-_36276930
|
0.971
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr11_+_45826640
|
0.964
|
NM_018389
|
SLC35C1
|
solute carrier family 35, member C1
|
|
chrX_-_142722869
|
0.960
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr5_-_16617085
|
0.948
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr10_+_124768401
|
0.945
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr10_-_70287249
|
0.942
|
NM_152707
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16
|
|
chrX_+_14903412
|
0.931
|
NM_001177475
|
MOSPD2
|
motile sperm domain containing 2
|
|
chr6_-_3445198
|
0.931
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr4_-_42658883
|
0.929
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr12_-_58026846
|
0.924
|
NM_001478
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1
|
|
chr3_-_133614421
|
0.922
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr14_+_66974124
|
0.922
|
NM_001024218
NM_020806
|
GPHN
|
gephyrin
|
|
chr1_+_110546569
|
0.917
|
NM_001242675
NM_001242676
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr8_+_56015013
|
0.914
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr12_-_104234968
|
0.911
|
NM_001031701
|
NT5DC3
|
5'-nucleotidase domain containing 3
|
|
chr8_+_67624652
|
0.901
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chrX_+_119738542
|
0.900
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr8_+_1449531
|
0.894
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr4_+_2061238
|
0.879
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr1_+_25870043
|
0.878
|
NM_015627
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1
|
|
chr2_+_45878912
|
0.877
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chrX_-_142722318
|
0.876
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr1_+_110543548
|
0.874
|
NM_001242674
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr11_-_88781039
|
0.868
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr5_-_74326723
|
0.868
|
NM_016591
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2
|
|
chr2_+_166150340
|
0.858
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr1_-_231560789
|
0.852
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr1_+_230202928
|
0.843
|
NM_004481
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2)
|
|
chr10_+_11047253
|
0.843
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr6_+_17600504
|
0.840
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr7_+_87257704
|
0.838
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr2_-_152955502
|
0.835
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr1_-_86043932
|
0.830
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr15_+_31619043
|
0.824
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr8_+_65492794
|
0.824
|
NM_152414
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr19_+_48958765
|
0.820
|
NM_013348
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr9_+_79056581
|
0.820
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr4_+_86396251
|
0.820
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr4_+_166128769
|
0.815
|
NM_007246
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr19_+_42817434
|
0.810
|
NM_173633
|
TMEM145
|
transmembrane protein 145
|
|
chr5_+_67588395
|
0.810
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr2_-_152955208
|
0.798
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr13_+_97874540
|
0.792
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr15_-_55563091
|
0.791
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr5_+_6448735
|
0.787
|
NM_001145161
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr19_-_33555757
|
0.787
|
NM_033103
|
RHPN2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr9_-_140115774
|
0.787
|
NM_031297
|
RNF208
|
ring finger protein 208
|
|
chr6_+_11094265
|
0.786
|
NM_001135575
|
C6orf228
|
chromosome 6 open reading frame 228
|
|
chr20_+_57766074
|
0.785
|
NM_178457
|
ZNF831
|
zinc finger protein 831
|
|
chr1_+_110527327
|
0.785
|
NM_001242673
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr6_-_91006460
|
0.780
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr7_+_114055048
|
0.777
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chrX_+_49832214
|
0.775
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr17_+_75446612
|
0.772
|
NM_001113496
|
SEPT9
|
septin 9
|
|
chr2_+_54952148
|
0.765
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr6_+_64281910
|
0.762
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr1_-_109940497
|
0.759
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr6_-_91006580
|
0.758
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_+_166326156
|
0.754
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr8_-_130951996
|
0.739
|
NM_016623
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr11_-_8680382
|
0.737
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr5_+_67586466
|
0.737
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chrX_+_14891526
|
0.736
|
NM_152581
|
MOSPD2
|
motile sperm domain containing 2
|
|
chr8_-_125740650
|
0.736
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr9_-_115480363
|
0.734
|
NM_021218
|
C9orf80
|
chromosome 9 open reading frame 80
|
|
chr2_-_175499275
|
0.731
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr2_+_32288579
|
0.731
|
NM_014946
NM_199436
|
SPAST
|
spastin
|
|
chr4_-_102268626
|
0.731
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr13_+_24734788
|
0.728
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chrX_-_48769234
|
0.727
|
NM_001042498
|
SLC35A2
|
solute carrier family 35 (UDP-galactose transporter), member A2
|
|
chr12_-_25403824
|
0.726
|
NM_004985
NM_033360
|
KRAS
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
|
|
chr2_-_44587812
|
0.724
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr19_+_19174792
|
0.721
|
NM_178526
|
SLC25A42
|
solute carrier family 25, member 42
|
|
chr5_+_89770680
|
0.718
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr16_-_52580805
|
0.718
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr9_+_79115548
|
0.718
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr10_+_75910908
|
0.717
|
NM_001202450
NM_006721
|
ADK
|
adenosine kinase
|
|
chr5_-_37835592
|
0.711
|
NM_199231
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr16_-_89007605
|
0.708
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr3_+_5229357
|
0.707
|
NM_014674
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr2_+_166428845
|
0.706
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr12_+_50135292
|
0.706
|
NM_003217
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr18_+_55102777
|
0.706
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr18_+_47088400
|
0.703
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr12_-_106533810
|
0.700
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|