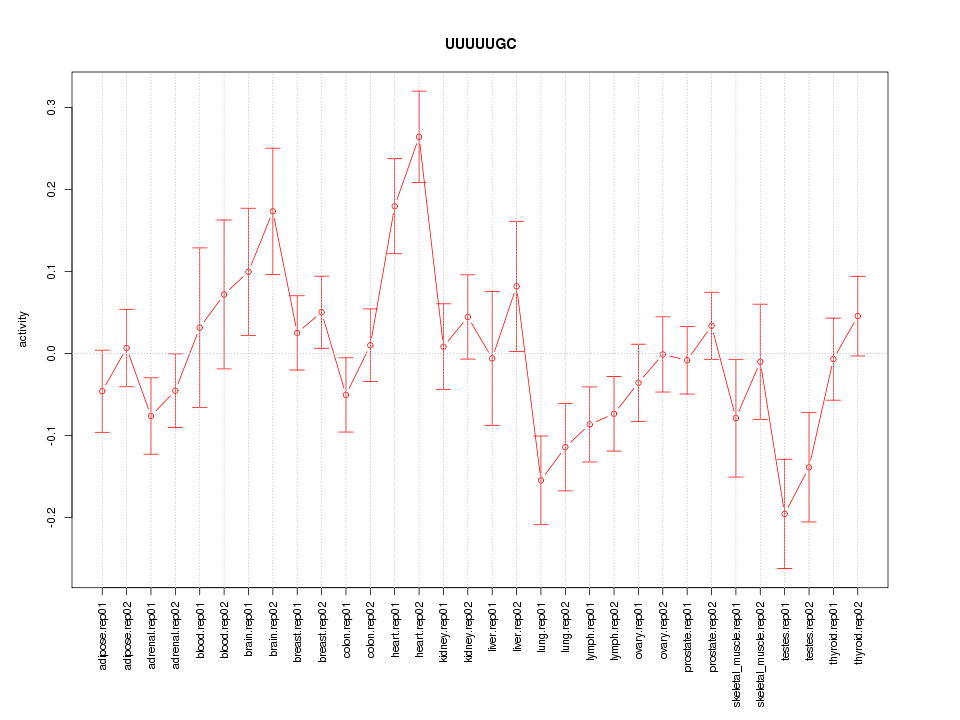
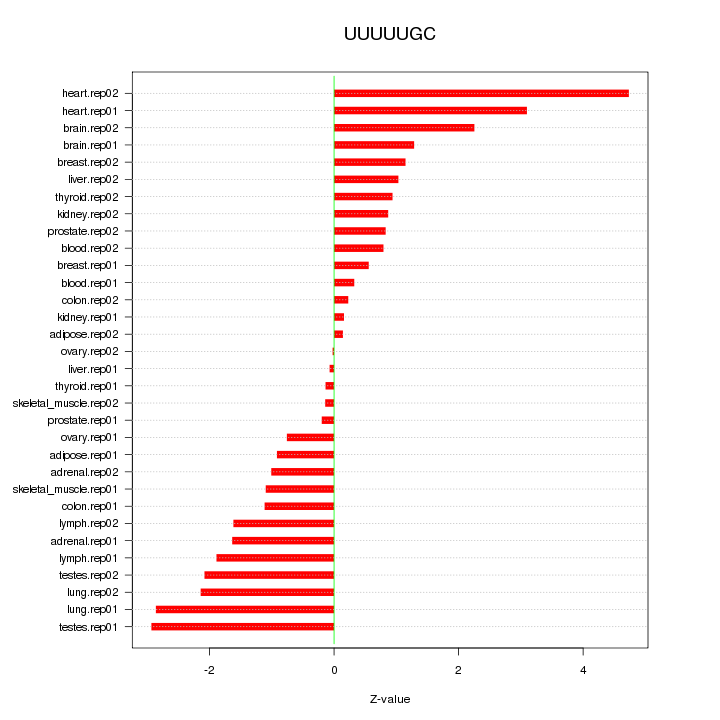
|
chr4_-_65275000
|
3.598
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr16_-_46782104
|
3.310
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr18_+_43304091
|
3.285
|
NM_001128588
NM_001146036
NM_015865
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr3_-_46608039
|
2.936
|
NM_024512
|
LRRC2
|
leucine rich repeat containing 2
|
|
chr3_-_192445344
|
2.914
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr3_-_192126837
|
2.855
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr18_+_43306917
|
2.681
|
NM_001146037
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr4_-_46995440
|
2.492
|
NM_001204266
NM_001204267
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr4_-_46996423
|
2.416
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr16_+_5008317
|
2.072
|
NM_014692
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae)
|
|
chr15_+_40697685
|
2.005
|
NM_001159508
NM_002225
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr1_+_237205701
|
1.832
|
NM_001035
|
RYR2
|
ryanodine receptor 2 (cardiac)
|
|
chr4_+_186064416
|
1.808
|
NM_001151
|
SLC25A4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4
|
|
chr4_-_89205887
|
1.662
|
NM_152542
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K
|
|
chr6_+_24495166
|
1.660
|
NM_001080
NM_170740
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1
|
|
chr7_-_16460946
|
1.649
|
NM_001101417
NM_001101426
|
ISPD
|
isoprenoid synthase domain containing
|
|
chr15_-_42749710
|
1.628
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr9_-_104198045
|
1.558
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr16_+_66968319
|
1.523
|
NM_003869
NM_198061
|
CES2
|
carboxylesterase 2
|
|
chr1_-_169555710
|
1.519
|
NM_000130
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr6_-_53530505
|
1.505
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr2_-_50574891
|
1.458
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr1_+_95558072
|
1.437
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr22_+_32439018
|
1.425
|
NM_000343
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1
|
|
chr1_-_193155723
|
1.405
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr5_+_130506635
|
1.404
|
NM_181705
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr18_+_32290195
|
1.367
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr12_+_119419299
|
1.348
|
NM_194286
|
SRRM4
|
serine/arginine repetitive matrix 4
|
|
chr1_+_24742244
|
1.341
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr2_+_207630111
|
1.334
|
NM_001136193
NM_001136194
NM_014929
|
FASTKD2
|
FAST kinase domains 2
|
|
chr9_+_112403067
|
1.323
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr7_-_25164902
|
1.317
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chrX_+_84258831
|
1.313
|
NM_198450
|
APOOL
|
apolipoprotein O-like
|
|
chr13_-_67804432
|
1.310
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr6_+_53883713
|
1.305
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr19_-_51920841
|
1.294
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr1_-_100715388
|
1.274
|
NM_001918
|
DBT
|
dihydrolipoamide branched chain transacylase E2
|
|
chr22_-_39240016
|
1.272
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr1_+_10003485
|
1.269
|
NM_022787
|
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1
|
|
chr11_-_35441501
|
1.264
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr3_+_38537749
|
1.223
|
NM_001145464
NM_005107
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like
|
|
chr11_-_19223516
|
1.203
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr4_+_184826427
|
1.191
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr3_-_69171536
|
1.177
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr7_+_86273219
|
1.176
|
NM_000840
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr10_-_113943467
|
1.172
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr3_-_62861012
|
1.172
|
NM_003716
NM_183393
NM_183394
|
CADPS
|
Ca++-dependent secretion activator
|
|
chr7_+_140774031
|
1.169
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr4_+_120056938
|
1.159
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr19_-_40562107
|
1.149
|
NM_001005851
|
ZNF780B
|
zinc finger protein 780B
|
|
chr2_-_175351755
|
1.148
|
NM_001033045
NM_152529
|
GPR155
|
G protein-coupled receptor 155
|
|
chr13_-_27334777
|
1.143
|
NM_005288
|
GPR12
|
G protein-coupled receptor 12
|
|
chr2_+_169312793
|
1.137
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr11_-_35441099
|
1.131
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr19_-_20150251
|
1.113
|
NM_033196
|
ZNF682
|
zinc finger protein 682
|
|
chr1_+_95582827
|
1.112
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr9_-_4726226
|
1.111
|
NM_001199856
|
AK3
|
adenylate kinase 3
|
|
chr3_-_184971815
|
1.102
|
NM_001166415
NM_001966
|
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase
|
|
chr10_+_12110921
|
1.101
|
NM_018706
|
DHTKD1
|
dehydrogenase E1 and transketolase domain containing 1
|
|
chr3_+_85775631
|
1.065
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr3_+_133464883
|
1.064
|
NM_001063
|
TF
|
transferrin
|
|
chr3_+_158991035
|
1.062
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr19_-_23578226
|
1.051
|
NM_003430
|
ZNF91
|
zinc finger protein 91
|
|
chr1_-_24438625
|
1.043
|
NM_152372
|
MYOM3
|
myomesin family, member 3
|
|
chr17_-_9862765
|
1.027
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr19_-_20150013
|
1.017
|
NM_001077349
|
ZNF682
|
zinc finger protein 682
|
|
chr4_+_81187741
|
0.996
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr9_+_74729510
|
0.987
|
NM_001242507
|
GDA
|
guanine deaminase
|
|
chr6_-_90348435
|
0.986
|
NM_020466
|
LYRM2
|
LYR motif containing 2
|
|
chr1_+_53662100
|
0.979
|
NM_000098
|
CPT2
|
carnitine palmitoyltransferase 2
|
|
chr8_-_74659942
|
0.975
|
NM_001164384
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr18_-_28681885
|
0.967
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chrX_+_72782961
|
0.962
|
NM_001039840
|
CHIC1
|
cysteine-rich hydrophobic domain 1
|
|
chr1_-_29557419
|
0.953
|
NM_001024732
NM_016011
|
MECR
|
mitochondrial trans-2-enoyl-CoA reductase
|
|
chr6_-_134373618
|
0.950
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr2_-_178937472
|
0.943
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr8_-_133459508
|
0.940
|
NM_001204824
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr1_+_160085519
|
0.937
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr8_+_94767071
|
0.928
|
NM_001142301
NM_153704
|
TMEM67
|
transmembrane protein 67
|
|
chrX_+_12885190
|
0.925
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chrX_+_154299674
|
0.921
|
NM_001018055
NM_001242640
NM_024332
|
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3
|
|
chr17_-_9940004
|
0.917
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr10_+_88428167
|
0.916
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr11_-_30607762
|
0.914
|
NM_001145399
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr6_-_105627857
|
0.914
|
NM_022361
|
POPDC3
|
popeye domain containing 3
|
|
chr9_+_108320382
|
0.907
|
NM_001079802
NM_001198963
NM_006731
|
FKTN
|
fukutin
|
|
chr7_-_99573637
|
0.906
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr3_+_159557649
|
0.905
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr7_+_56032065
|
0.905
|
NM_001202469
NM_001483
|
GBAS
|
glioblastoma amplified sequence
|
|
chr2_-_51259536
|
0.904
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chrX_-_63005212
|
0.902
|
NM_001173479
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr2_-_208890207
|
0.901
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr6_-_46293628
|
0.897
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr6_-_35696359
|
0.889
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr19_-_53238290
|
0.879
|
NM_001161501
NM_001161499
NM_001161500
|
ZNF611
|
zinc finger protein 611
|
|
chr18_-_24442534
|
0.877
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr19_+_48774653
|
0.876
|
NM_153608
|
ZNF114
|
zinc finger protein 114
|
|
chr3_+_121286777
|
0.874
|
NM_001012659
|
ARGFX
|
arginine-fifty homeobox
|
|
chr3_-_122746572
|
0.868
|
NM_001031702
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B
|
|
chr15_-_66545994
|
0.867
|
NM_032445
|
MEGF11
|
multiple EGF-like-domains 11
|
|
chrX_+_12156584
|
0.862
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr5_-_78809658
|
0.860
|
NM_004272
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr4_+_154074269
|
0.859
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr11_+_28129797
|
0.857
|
NM_001113528
NM_152636
|
METTL15
|
methyltransferase like 15
|
|
chrX_-_62975005
|
0.854
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr4_-_166034003
|
0.852
|
NM_001100389
|
TMEM192
|
transmembrane protein 192
|
|
chr7_-_139477437
|
0.848
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr3_-_172428773
|
0.844
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr5_-_16617085
|
0.843
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr19_-_22193720
|
0.841
|
NM_007153
|
ZNF208
|
zinc finger protein 208
|
|
chr2_+_187464931
|
0.840
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr5_-_125930604
|
0.839
|
NM_001182
NM_001201377
NM_001202404
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr11_-_133826879
|
0.837
|
NM_014987
|
IGSF9B
|
immunoglobulin superfamily, member 9B
|
|
chr3_+_179370779
|
0.834
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr6_+_96463844
|
0.833
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr3_+_101498028
|
0.832
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr9_-_4741267
|
0.830
|
NM_001199852
NM_016282
|
AK3
|
adenylate kinase 3
|
|
chrX_-_77041678
|
0.829
|
NM_000489
NM_138270
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr17_+_27369917
|
0.829
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chr12_+_113229548
|
0.827
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr11_-_134281725
|
0.827
|
NM_018644
NM_054025
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chr6_-_35656677
|
0.823
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr15_-_90437616
|
0.823
|
NM_005829
|
AP3S2
|
adaptor-related protein complex 3, sigma 2 subunit
|
|
chr9_-_4741085
|
0.822
|
NM_001199854
|
AK3
|
adenylate kinase 3
|
|
chr8_-_97247757
|
0.821
|
NM_001199975
NM_006294
|
UQCRB
|
ubiquinol-cytochrome c reductase binding protein
|
|
chr19_-_39523132
|
0.818
|
NM_178820
|
FBXO27
|
F-box protein 27
|
|
chr1_+_10270680
|
0.817
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr5_-_131347249
|
0.814
|
NM_001009185
NM_001205247
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr1_+_65730423
|
0.813
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr2_-_139537803
|
0.812
|
NM_007226
|
NXPH2
|
neurexophilin 2
|
|
chr3_+_101498285
|
0.810
|
NM_001134456
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr19_-_11591439
|
0.810
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr1_-_112298418
|
0.805
|
NM_198926
|
FAM212B
|
family with sequence similarity 212, member B
|
|
chr13_+_103451398
|
0.803
|
NM_001159596
NM_017693
|
BIVM
|
basic, immunoglobulin-like variable motif containing
|
|
chr17_-_9929567
|
0.803
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chrX_+_37545008
|
0.801
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr7_-_14880954
|
0.801
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr1_-_205744543
|
0.801
|
NM_001135662
NM_001135663
NM_001135664
NM_003929
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1
|
|
chr18_-_24445652
|
0.801
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr16_+_48278177
|
0.799
|
NM_031490
|
LONP2
|
lon peptidase 2, peroxisomal
|
|
chr14_+_24584467
|
0.797
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr5_-_10308167
|
0.794
|
NM_138809
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|
|
chr16_-_12897693
|
0.791
|
NM_001099455
NM_018340
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr13_-_102068705
|
0.791
|
NM_052867
|
NALCN
|
sodium leak channel, non-selective
|
|
chr6_-_163148800
|
0.788
|
NM_004562
NM_013987
NM_013988
|
PARK2
|
parkinson protein 2, E3 ubiquitin protein ligase (parkin)
|
|
chr5_-_16509106
|
0.786
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr20_+_58508818
|
0.783
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr2_-_44586854
|
0.782
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr15_+_62939509
|
0.781
|
NM_015059
|
TLN2
|
talin 2
|
|
chr10_-_75410778
|
0.780
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr2_-_178753464
|
0.777
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chr4_+_6784453
|
0.775
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr17_-_10101854
|
0.767
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr4_+_154125589
|
0.765
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_-_44587812
|
0.764
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr19_-_44143943
|
0.760
|
NM_145296
|
CADM4
|
cell adhesion molecule 4
|
|
chr1_-_112281864
|
0.760
|
NM_019099
|
FAM212B
|
family with sequence similarity 212, member B
|
|
chr2_-_44588622
|
0.759
|
NM_001171606
NM_001171613
|
PREPL
|
prolyl endopeptidase-like
|
|
chr3_-_132378492
|
0.756
|
NM_032169
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11
|
|
chr3_-_74570262
|
0.755
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chrX_-_19533378
|
0.754
|
NM_001001671
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15
|
|
chr19_+_54466289
|
0.752
|
NM_031895
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8
|
|
chr10_-_91403630
|
0.749
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr17_+_33474835
|
0.748
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr8_-_110986923
|
0.746
|
NM_014379
|
KCNV1
|
potassium channel, subfamily V, member 1
|
|
chr4_-_71705613
|
0.744
|
NM_002092
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr13_+_80055257
|
0.742
|
NM_001161407
NM_019080
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr9_-_4742042
|
0.740
|
NM_001199853
|
AK3
|
adenylate kinase 3
|
|
chr11_+_44785975
|
0.738
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr17_-_30669133
|
0.738
|
NM_022344
|
C17orf75
|
chromosome 17 open reading frame 75
|
|
chr9_+_74764266
|
0.736
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr11_-_133402227
|
0.735
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr2_-_178973065
|
0.734
|
NM_001077197
|
PDE11A
|
phosphodiesterase 11A
|
|
chr11_+_111895537
|
0.733
|
NM_001931
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr1_+_45965855
|
0.732
|
NM_015506
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria
|
|
chr11_-_62559423
|
0.727
|
NM_001080501
|
TMEM223
|
transmembrane protein 223
|
|
chr10_-_92681025
|
0.726
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr9_-_131084496
|
0.724
|
NM_015679
|
TRUB2
|
TruB pseudouridine (psi) synthase homolog 2 (E. coli)
|
|
chr2_+_173420776
|
0.719
|
NM_002610
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr3_+_85008031
|
0.718
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr5_-_96478502
|
0.717
|
NM_153234
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr14_-_50778788
|
0.715
|
NM_024884
|
L2HGDH
|
L-2-hydroxyglutarate dehydrogenase
|
|
chr13_+_21750371
|
0.714
|
NM_024026
|
MRP63
|
mitochondrial ribosomal protein 63
|
|
chr3_-_178789597
|
0.713
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr6_+_153071931
|
0.713
|
NM_003381
NM_194435
|
VIP
|
vasoactive intestinal peptide
|
|
chr6_+_24126349
|
0.712
|
NM_080723
|
NRSN1
|
neurensin 1
|
|
chr3_+_135741576
|
0.711
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr10_-_1779669
|
0.710
|
NM_018702
|
ADARB2
|
adenosine deaminase, RNA-specific, B2
|
|
chr9_+_71650478
|
0.708
|
NM_000144
NM_001161706
NM_181425
|
FXN
|
frataxin
|
|
chr7_-_5553305
|
0.705
|
NM_024963
|
FBXL18
|
F-box and leucine-rich repeat protein 18
|
|
chr8_-_87520981
|
0.702
|
NM_016033
|
FAM82B
|
family with sequence similarity 82, member B
|
|
chr20_+_24449834
|
0.702
|
NM_024893
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr4_-_71705188
|
0.701
|
NM_001098477
|
GRSF1
|
G-rich RNA sequence binding factor 1
|
|
chr10_+_104629209
|
0.701
|
NM_020682
|
AS3MT
|
arsenic (+3 oxidation state) methyltransferase
|
|
chr11_+_34937676
|
0.700
|
NM_001135024
NM_001166158
NM_003477
|
PDHX
|
pyruvate dehydrogenase complex, component X
|
|
chr3_-_12586962
|
0.699
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr10_-_75415748
|
0.698
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr6_-_18155283
|
0.696
|
NM_000367
|
TPMT
|
thiopurine S-methyltransferase
|
|
chr13_-_50510410
|
0.694
|
NM_001127482
NM_020456
|
SPRYD7
|
SPRY domain containing 7
|
|
chrX_-_142723921
|
0.693
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|