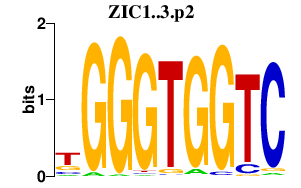
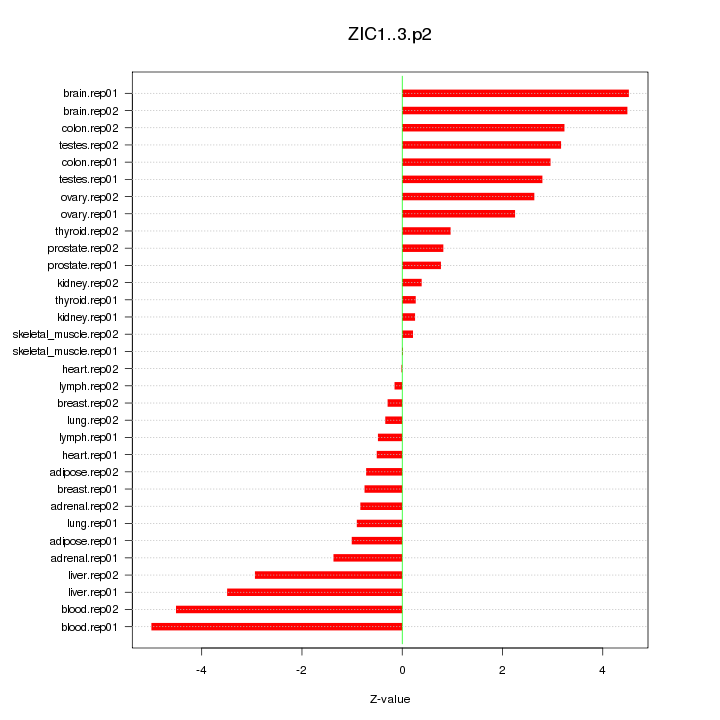
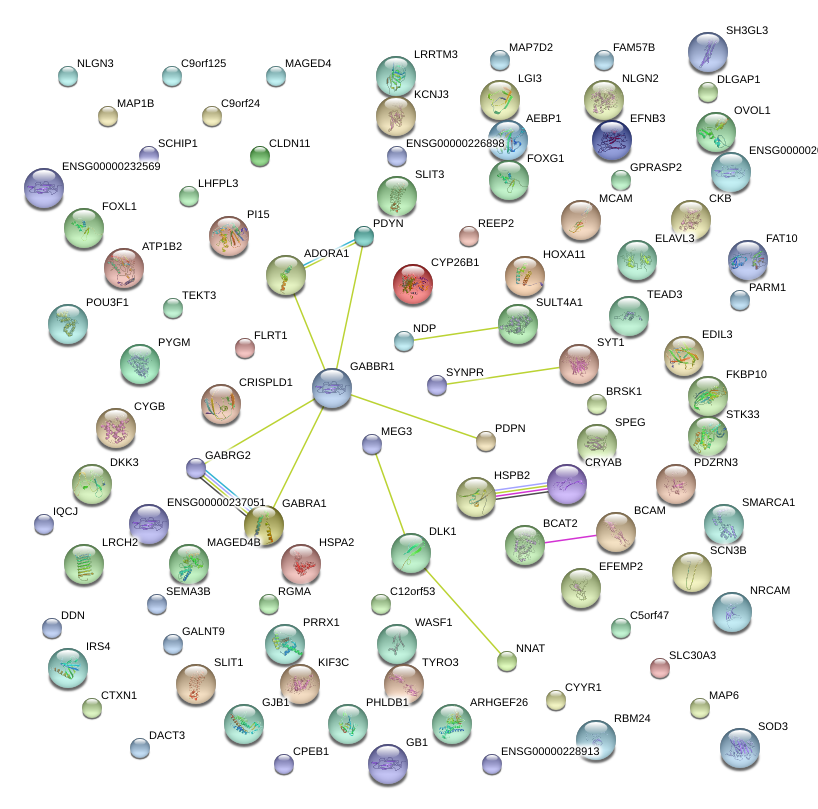
|
chr3_-_73673978
|
10.832
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr11_-_75379414
|
8.064
|
NM_033063
NM_207577
|
MAP6
|
microtubule-associated protein 6
|
|
chr8_-_22014263
|
7.452
|
NM_139278
|
LGI3
|
leucine-rich repeat LGI family, member 3
|
|
chr11_-_75379651
|
6.905
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr22_-_44258210
|
6.805
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr14_+_29236882
|
6.783
|
|
FOXG1
|
forkhead box G1
|
|
chr14_-_103988829
|
6.683
|
|
CKB
|
creatine kinase, brain
|
|
chr3_+_153839136
|
6.624
|
NM_001251963
NM_015595
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr11_-_111782447
|
6.506
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr19_-_47164289
|
6.281
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr11_-_75379113
|
6.078
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr12_-_49392909
|
5.982
|
NM_015086
|
DDN
|
dendrin
|
|
chr1_-_38512446
|
5.939
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr5_+_71403040
|
5.721
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_-_168727715
|
5.418
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr3_+_63263814
|
5.412
|
NM_001130003
|
SYNPR
|
synaptoporin
|
|
chrX_+_70364680
|
5.311
|
NM_001166660
NM_018977
NM_181303
|
NLGN3
|
neuroligin 3
|
|
chr3_+_170136652
|
5.207
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr8_+_75736771
|
5.193
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr11_+_111783476
|
5.163
|
|
HSPB2-C11orf52
HSPB2
|
HSPB2-C11orf52 readthrough (non-protein coding)
heat shock 27kDa protein 2
|
|
chrX_+_70364707
|
5.127
|
|
NLGN3
|
neuroligin 3
|
|
chr15_+_84116172
|
5.079
|
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr7_-_27224762
|
5.015
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr20_+_36149606
|
4.921
|
NM_005386
NM_181689
|
NNAT
|
neuronatin
|
|
chr11_-_12030539
|
4.845
|
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr14_+_101292451
|
4.735
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr14_+_29236171
|
4.665
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr17_-_74533623
|
4.664
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr5_+_173416161
|
4.624
|
NM_001144954
|
C5orf47
|
chromosome 5 open reading frame 47
|
|
chr10_+_68685791
|
4.608
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr11_-_123525300
|
4.599
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chrX_-_43832920
|
4.562
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr19_+_55795766
|
4.474
|
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr5_+_137774632
|
4.462
|
NM_016606
|
REEP2
|
receptor accessory protein 2
|
|
chr11_+_111783459
|
4.458
|
NM_001541
|
HSPB2-C11orf52
HSPB2
|
HSPB2-C11orf52 readthrough (non-protein coding)
heat shock 27kDa protein 2
|
|
chr16_+_86612114
|
4.233
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr15_+_84115979
|
4.231
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr14_-_103989195
|
4.226
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr7_+_103969103
|
4.210
|
NM_199000
|
LHFPL3
|
lipoma HMGIC fusion partner-like 3
|
|
chrX_+_101967103
|
4.053
|
NM_001004051
NM_001184874
NM_001184875
NM_001184876
NM_138437
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr14_-_103989112
|
4.018
|
|
CKB
|
creatine kinase, brain
|
|
chr11_-_12030622
|
4.006
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr11_-_12030128
|
3.989
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chrX_-_114468604
|
3.971
|
NM_001243963
NM_020871
|
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2
|
|
chr12_+_79258588
|
3.916
|
|
SYT1
|
synaptotagmin I
|
|
chr17_+_7608413
|
3.881
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr15_+_41851153
|
3.869
|
NM_006293
|
TYRO3
|
TYRO3 protein tyrosine kinase
|
|
chr11_-_64527369
|
3.862
|
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr3_+_64670779
|
3.839
|
|
ADAMTS9-AS2
|
ADAMTS9 antisense RNA 2 (non-protein coding)
|
|
chrX_-_20135015
|
3.825
|
NM_001168465
NM_001168466
NM_152780
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr11_-_8615701
|
3.761
|
|
STK33
|
serine/threonine kinase 33
|
|
chr6_-_29595934
|
3.728
|
NM_021903
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr5_-_83680191
|
3.686
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr2_-_26205327
|
3.672
|
|
KIF3C
|
kinesin family member 3C
|
|
chr11_-_119187811
|
3.629
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr2_+_155555092
|
3.610
|
NM_002239
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3
|
|
chrX_+_70364728
|
3.602
|
|
NLGN3
|
neuroligin 3
|
|
chr3_+_159481880
|
3.583
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr22_+_36960325
|
3.579
|
|
|
|
|
chr5_+_161494647
|
3.564
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr12_-_6809965
|
3.556
|
NM_001244014
NM_153685
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
chrX_-_13956445
|
3.538
|
|
|
|
|
chr19_+_55795533
|
3.529
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr1_+_170633312
|
3.511
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr2_-_27485774
|
3.488
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr17_+_39968961
|
3.458
|
NM_021939
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr12_+_79258517
|
3.436
|
|
SYT1
|
synaptotagmin I
|
|
chrX_-_51812249
|
3.427
|
NM_001098800
NM_001242362
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr11_-_8615412
|
3.376
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr12_+_79258794
|
3.365
|
|
SYT1
|
synaptotagmin I
|
|
chr9_-_104249333
|
3.350
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr21_-_27945267
|
3.341
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr6_-_35464716
|
3.334
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr7_+_44144002
|
3.307
|
|
AEBP1
|
AE binding protein 1
|
|
chr17_+_7311501
|
3.300
|
NM_020795
|
NLGN2
|
neuroligin 2
|
|
chr17_-_15244898
|
3.293
|
NM_031898
|
TEKT3
|
tektin 3
|
|
chr2_+_220325533
|
3.225
|
NM_001173476
|
SPEG
|
SPEG complex locus
|
|
chrX_-_107979606
|
3.209
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr3_+_50306578
|
3.187
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr5_+_161275541
|
3.177
|
NM_001127645
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr11_-_65640199
|
3.139
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr1_+_203096835
|
3.115
|
NM_000674
NM_001048230
|
ADORA1
|
adenosine A1 receptor
|
|
chr19_-_11591439
|
3.112
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr18_-_3845295
|
3.111
|
NM_001003809
NM_001242765
NM_001242766
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr8_+_75896707
|
3.103
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr20_-_1974701
|
3.076
|
NM_001190898
NM_001190899
NM_024411
|
PDYN
|
prodynorphin
|
|
chr1_+_13910251
|
3.074
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr19_+_45312294
|
3.074
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr11_+_118478305
|
3.070
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr7_-_108096693
|
3.056
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr16_-_30042123
|
3.047
|
NM_031478
|
FAM57B
|
family with sequence similarity 57, member B
|
|
chr12_-_132690572
|
3.036
|
NM_021808
|
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9)
|
|
chr3_+_50306305
|
3.015
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr4_+_24797203
|
3.009
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr14_+_101193313
|
3.008
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr15_-_93616891
|
2.992
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chrX_+_70443055
|
2.984
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr7_+_44143945
|
2.972
|
NM_001129
|
AEBP1
|
AE binding protein 1
|
|
chrX_+_49047887
|
2.969
|
|
|
|
|
chr19_-_7990974
|
2.965
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr6_+_17281773
|
2.936
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr14_+_101292483
|
2.927
|
|
|
|
|
chr11_-_8615745
|
2.919
|
|
STK33
|
serine/threonine kinase 33
|
|
chrX_-_128657453
|
2.894
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr6_-_29595746
|
2.885
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr11_+_63871240
|
2.882
|
NM_013280
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1
|
|
chr9_-_34397786
|
2.881
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr6_-_110500803
|
2.873
|
|
WASF1
|
WAS protein family, member 1
|
|
chr14_+_101193190
|
2.846
|
NM_003836
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr11_-_123524881
|
2.830
|
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr4_+_75858205
|
2.822
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr11_+_65554504
|
2.815
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr17_+_7554415
|
2.795
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chrX_+_101967290
|
2.795
|
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr15_-_83316621
|
2.795
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr14_+_65007418
|
2.792
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr2_-_72374919
|
2.791
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chrX_-_30327479
|
2.782
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr9_-_122131579
|
2.770
|
NM_014618
|
DBC1
|
deleted in bladder cancer 1
|
|
chr4_+_24797084
|
2.760
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr2_+_220283098
|
2.757
|
NM_001927
|
DES
|
desmin
|
|
chr9_-_73483933
|
2.733
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr7_+_119913688
|
2.731
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chrX_+_102000906
|
2.729
|
NM_001142529
NM_001142530
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr1_+_236849753
|
2.728
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr17_-_50237342
|
2.716
|
NM_001082534
|
CA10
|
carbonic anhydrase X
|
|
chr5_+_140855604
|
2.697
|
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr9_-_138391696
|
2.691
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr13_-_36871968
|
2.687
|
NM_001198910
NM_001144981
NM_001144982
NM_001144983
NM_001144984
NM_001144985
NM_001144986
NM_001198908
|
CCDC169-SOHLH2
CCDC169
|
CCDC169-SOHLH2 readthrough
coiled-coil domain containing 169
|
|
chr22_-_28197469
|
2.662
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr5_-_134914447
|
2.660
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr2_+_16081945
|
2.657
|
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr6_+_163149001
|
2.657
|
NM_001080379
|
PACRG
|
PARK2 co-regulated
|
|
chr3_-_12200850
|
2.656
|
NM_003256
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chrX_+_90689596
|
2.656
|
NM_080832
|
PABPC5
|
poly(A) binding protein, cytoplasmic 5
|
|
chrX_+_51927918
|
2.656
|
NM_001098800
NM_001242362
NM_030801
NM_177535
NM_177537
|
MAGED4
MAGED4B
|
melanoma antigen family D, 4
melanoma antigen family D, 4B
|
|
chr6_+_41606193
|
2.653
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr17_+_47653474
|
2.649
|
|
NXPH3
|
neurexophilin 3
|
|
chr12_+_58003962
|
2.641
|
NM_001111270
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr11_+_66045645
|
2.620
|
NM_182553
|
CNIH2
|
cornichon homolog 2 (Drosophila)
|
|
chr10_-_119301750
|
2.614
|
|
EMX2OS
|
EMX2 opposite strand/antisense RNA (non-protein coding)
|
|
chr16_-_74808699
|
2.612
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr9_+_103235634
|
2.611
|
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr1_+_202091971
|
2.586
|
NM_004767
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chr22_-_42765179
|
2.584
|
|
|
|
|
chr18_-_31803514
|
2.582
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr3_-_195306274
|
2.581
|
|
APOD
|
apolipoprotein D
|
|
chr14_+_85996512
|
2.568
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr18_-_31803168
|
2.552
|
|
NOL4
|
nucleolar protein 4
|
|
chr12_-_53448167
|
2.549
|
|
LOC283335
|
uncharacterized LOC283335
|
|
chr1_-_45308603
|
2.538
|
NM_001166292
NM_003738
|
PTCH2
|
patched 2
|
|
chr14_+_94393687
|
2.534
|
NM_001207074
NM_001207073
NM_001207072
|
FAM181A
|
family with sequence similarity 181, member A
|
|
chr2_-_193059599
|
2.534
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chrX_-_128657381
|
2.509
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr3_+_153838791
|
2.500
|
NM_001251962
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr6_-_6007632
|
2.497
|
NM_016588
|
NRN1
|
neuritin 1
|
|
chr3_-_9595412
|
2.471
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr16_-_74808474
|
2.470
|
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr2_-_213403280
|
2.465
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr6_+_26204779
|
2.463
|
NM_003545
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr1_+_42846427
|
2.463
|
NM_173642
|
RIMKLA
|
ribosomal modification protein rimK-like family member A
|
|
chr19_-_44306525
|
2.461
|
NM_001031749
|
LYPD5
|
LY6/PLAUR domain containing 5
|
|
chr3_+_6902812
|
2.458
|
|
GRM7
|
glutamate receptor, metabotropic 7
|
|
chr15_-_93617091
|
2.454
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr19_-_49220213
|
2.437
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr16_-_325867
|
2.433
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr13_+_31480311
|
2.427
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr14_+_101295409
|
2.418
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr19_-_51071279
|
2.414
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr3_+_181430075
|
2.393
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr12_-_57410275
|
2.381
|
NM_001178054
NM_013251
|
TAC3
|
tachykinin 3
|
|
chr18_+_21452981
|
2.367
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr17_-_7197866
|
2.366
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr12_+_117176085
|
2.362
|
NM_001109903
NM_032814
|
RNFT2
|
ring finger protein, transmembrane 2
|
|
chr17_+_30593194
|
2.354
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr4_-_90758126
|
2.350
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr17_-_7197811
|
2.343
|
|
YBX2
|
Y box binding protein 2
|
|
chr2_+_133174146
|
2.336
|
NM_001508
|
GPR39
|
G protein-coupled receptor 39
|
|
chr16_+_55512801
|
2.329
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr10_+_119302709
|
2.323
|
|
|
|
|
chr4_+_4861371
|
2.319
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr15_+_74466887
|
2.315
|
NM_201526
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr9_-_113800243
|
2.312
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr1_+_220701547
|
2.298
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr13_-_36871882
|
2.291
|
|
CCDC169
|
coiled-coil domain containing 169
|
|
chrX_-_128657440
|
2.286
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr11_+_65554566
|
2.268
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr8_-_21988518
|
2.268
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr16_+_76343734
|
2.262
|
NM_138994
|
CNTNAP4
|
contactin associated protein-like 4
|
|
chr2_+_149633173
|
2.257
|
|
|
|
|
chr1_+_110693131
|
2.226
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr5_+_137774897
|
2.215
|
|
REEP2
|
receptor accessory protein 2
|
|
chr7_+_90032647
|
2.214
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr13_-_53422773
|
2.213
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr11_+_131780537
|
2.199
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr1_-_156217717
|
2.198
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr11_-_119252364
|
2.187
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr17_+_7554711
|
2.183
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr2_-_105467912
|
2.183
|
|
LOC100506421
|
uncharacterized LOC100506421
|
|
chr1_-_40105296
|
2.180
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|