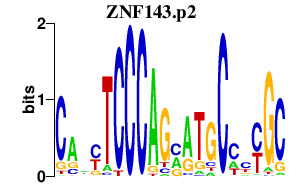
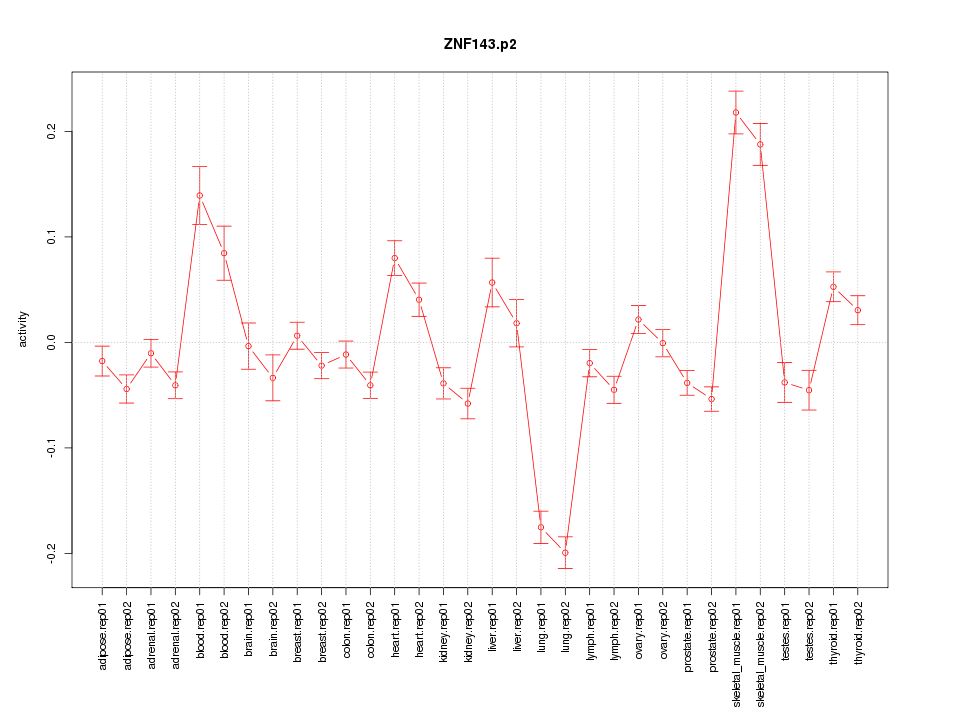
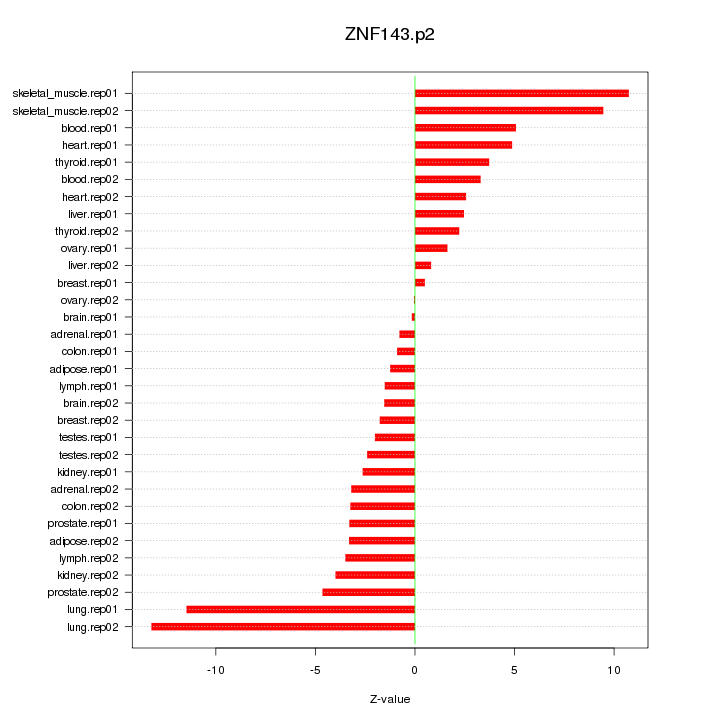
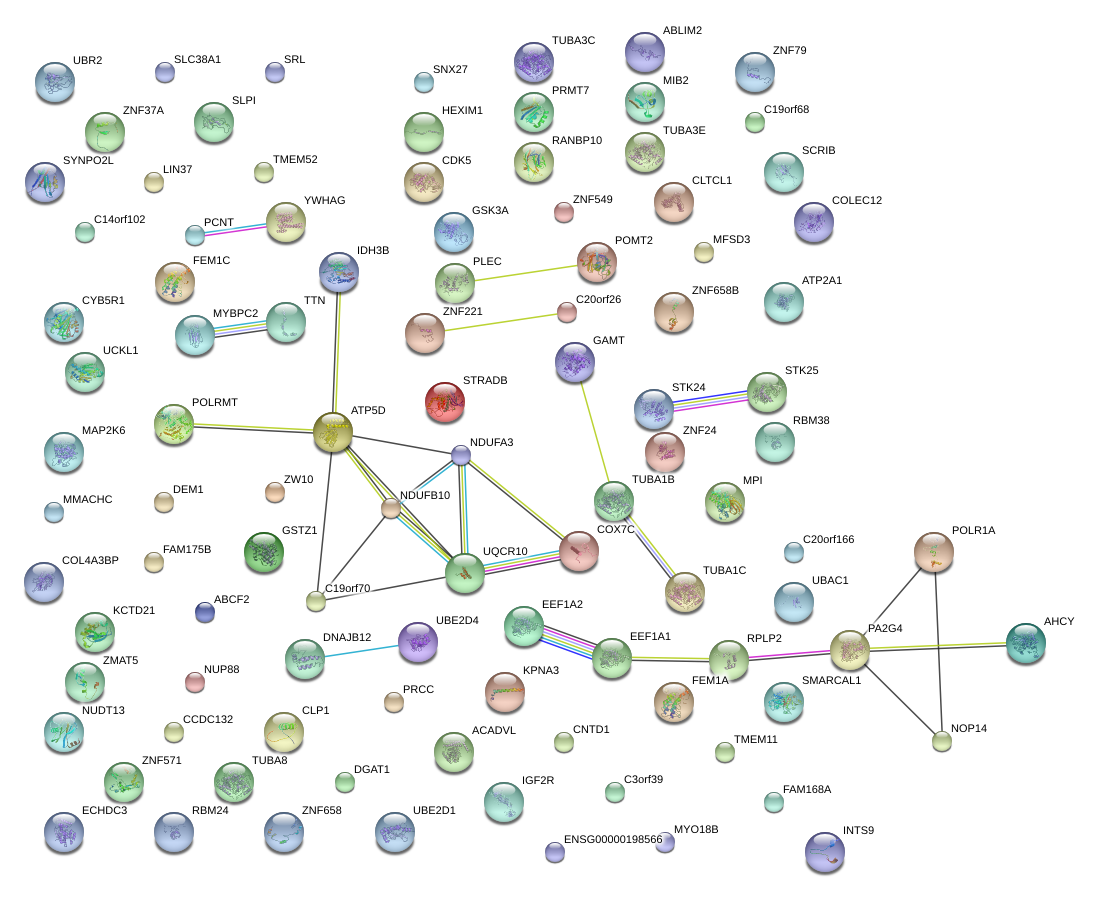
|
chr19_+_4791756
|
8.212
|
|
FEM1A
|
fem-1 homolog a (C. elegans)
|
|
chr19_+_4791727
|
7.148
|
NM_018708
|
FEM1A
|
fem-1 homolog a (C. elegans)
|
|
chr4_-_8160435
|
7.131
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr2_-_179672056
|
6.576
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr6_+_42531767
|
6.363
|
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr20_-_62130390
|
5.880
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr4_-_2965147
|
5.466
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr21_+_47744466
|
5.117
|
|
PCNT
|
pericentrin
|
|
chr2_-_242447449
|
5.104
|
|
STK25
|
serine/threonine kinase 25
|
|
chr19_+_48673936
|
4.906
|
|
C19orf68
|
chromosome 19 open reading frame 68
|
|
chr19_-_5680503
|
4.878
|
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr19_+_50936159
|
4.772
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr20_+_55966440
|
4.715
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr20_+_20033157
|
4.663
|
NM_015585
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr1_-_1850704
|
4.628
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr6_+_42531736
|
4.558
|
NM_001184801
NM_015255
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr10_+_74870209
|
4.457
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr22_+_26138110
|
4.417
|
NM_032608
|
MYO18B
|
myosin XVIIIB
|
|
chr9_-_40792062
|
4.416
|
NM_033160
|
ZNF658
|
zinc finger protein 658
|
|
chr22_+_30163351
|
4.329
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr2_-_242448000
|
4.293
|
NM_006374
|
STK25
|
serine/threonine kinase 25
|
|
chr19_-_42746743
|
4.272
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr22_+_26138168
|
4.271
|
|
MYO18B
|
myosin XVIIIB
|
|
chr4_-_2965036
|
4.216
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr4_-_2965091
|
4.213
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr19_-_42746678
|
4.192
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr6_+_42531951
|
4.162
|
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr14_-_90798253
|
4.159
|
NM_017970
|
C14orf102
|
chromosome 14 open reading frame 102
|
|
chr20_+_61147659
|
4.069
|
NM_178463
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr16_+_28891234
|
4.048
|
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr4_-_2965111
|
4.042
|
NM_003703
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr1_+_156024516
|
3.999
|
NM_001145264
NM_014017
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2
|
|
chr7_-_150754934
|
3.979
|
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr19_+_4791757
|
3.878
|
|
FEM1A
|
fem-1 homolog a (C. elegans)
|
|
chr14_+_77787229
|
3.844
|
NM_145870
NM_145871
|
GSTZ1
|
glutathione transferase zeta 1
|
|
chr16_+_2009516
|
3.791
|
NM_004548
|
NDUFB10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa
|
|
chr10_+_38383218
|
3.758
|
NM_001007094
NM_001178101
NM_003421
|
ZNF37A
|
zinc finger protein 37A
|
|
chr10_-_75415748
|
3.755
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr1_+_45965855
|
3.746
|
NM_015506
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria
|
|
chr19_-_1401474
|
3.738
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr13_-_50367027
|
3.731
|
NM_002267
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr22_-_30162990
|
3.710
|
|
|
|
|
chr22_+_18593399
|
3.650
|
NM_001193414
NM_018943
|
TUBA8
|
tubulin, alpha 8
|
|
chr1_+_156737315
|
3.646
|
|
PRCC
|
papillary renal cell carcinoma (translocation-associated)
|
|
chr17_+_67410825
|
3.646
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr1_-_202936333
|
3.638
|
NM_016243
|
CYB5R1
|
cytochrome b5 reductase 1
|
|
chr10_+_11784355
|
3.593
|
NM_024693
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|
|
chr21_+_47744032
|
3.592
|
NM_006031
|
PCNT
|
pericentrin
|
|
chr2_-_86332930
|
3.591
|
|
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa
|
|
chr11_-_113644432
|
3.590
|
|
ZW10
|
ZW10, kinetochore associated, homolog (Drosophila)
|
|
chr11_-_113644392
|
3.562
|
|
ZW10
|
ZW10, kinetochore associated, homolog (Drosophila)
|
|
chr22_-_30162933
|
3.557
|
NM_001003692
NM_019103
|
ZMAT5
|
zinc finger, matrin-type 5
|
|
chr10_+_126490353
|
3.520
|
NM_032182
|
FAM175B
|
family with sequence similarity 175, member B
|
|
chr11_-_113644476
|
3.445
|
NM_004724
|
ZW10
|
ZW10, kinetochore associated, homolog (Drosophila)
|
|
chr22_-_19279183
|
3.403
|
NM_001835
NM_007098
|
CLTCL1
|
clathrin, heavy chain-like 1
|
|
chr15_+_75182351
|
3.369
|
NM_002435
|
MPI
|
mannose phosphate isomerase
|
|
chr19_-_42746723
|
3.332
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr19_+_1241767
|
3.322
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr17_+_7123246
|
3.286
|
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr7_+_92861652
|
3.246
|
NM_017667
NM_024553
|
CCDC132
|
coiled-coil domain containing 132
|
|
chr7_-_150924220
|
3.232
|
NM_005692
NM_007189
|
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2
|
|
chr9_-_138853007
|
3.226
|
|
UBAC1
|
UBA domain containing 1
|
|
chr19_+_44455379
|
3.205
|
NM_013359
|
ZNF221
|
zinc finger protein 221
|
|
chr2_-_242447919
|
3.201
|
|
STK25
|
serine/threonine kinase 25
|
|
chr8_-_145013628
|
3.201
|
NM_201384
|
PLEC
|
plectin
|
|
chr19_+_58038926
|
3.168
|
|
ZNF549
|
zinc finger protein 549
|
|
chr19_+_1241742
|
3.153
|
NM_001001975
NM_001687
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr2_+_217277136
|
3.152
|
NM_014140
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1
|
|
chr9_-_138853146
|
3.131
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr19_+_1241811
|
3.123
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr14_-_77787023
|
3.104
|
NM_013382
|
POMT2
|
protein-O-mannosyltransferase 2
|
|
chr17_-_5322982
|
3.103
|
NM_002532
|
NUP88
|
nucleoporin 88kDa
|
|
chr16_-_67840464
|
3.094
|
NM_020850
|
RANBP10
|
RAN binding protein 10
|
|
chr19_-_42746544
|
3.071
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr7_-_150754979
|
3.062
|
NM_001164410
NM_004935
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr8_-_28747422
|
3.056
|
NM_001172562
|
INTS9
|
integrator complex subunit 9
|
|
chr20_-_32891075
|
3.048
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr1_+_156737273
|
3.047
|
NM_005973
|
PRCC
|
papillary renal cell carcinoma (translocation-associated)
|
|
chr17_-_21117647
|
3.015
|
|
TMEM11
|
transmembrane protein 11
|
|
chr5_+_85913762
|
3.003
|
NM_001867
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr19_-_633526
|
2.998
|
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr10_-_74114655
|
2.960
|
|
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12
|
|
chr16_+_68344868
|
2.942
|
NM_001184824
NM_019023
|
PRMT7
|
protein arginine methyltransferase 7
|
|
chr11_-_77899640
|
2.916
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr19_+_54606141
|
2.913
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr1_+_1551204
|
2.889
|
NM_001170689
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr8_+_145734445
|
2.859
|
NM_138431
|
MFSD3
|
major facilitator superfamily domain containing 3
|
|
chr8_-_144897541
|
2.822
|
NM_015356
NM_182706
|
SCRIB
|
scribbled homolog (Drosophila)
|
|
chr19_+_1241766
|
2.817
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr7_-_75988124
|
2.808
|
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr20_-_62587718
|
2.801
|
NM_017859
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr9_+_130186652
|
2.800
|
NM_007135
|
ZNF79
|
zinc finger protein 79
|
|
chr10_+_60094712
|
2.783
|
NM_001204880
NM_003338
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1
|
|
chr17_+_40950809
|
2.783
|
NM_173478
|
CNTD1
|
cyclin N-terminal domain containing 1
|
|
chr17_+_7123149
|
2.741
|
NM_000018
NM_001033859
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr18_-_32924381
|
2.728
|
|
ZNF24
|
zinc finger protein 24
|
|
chr3_-_43147470
|
2.715
|
NM_032806
|
C3orf39
|
chromosome 3 open reading frame 39
|
|
chr16_-_4292052
|
2.707
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr11_+_809918
|
2.697
|
NM_001004
|
RPLP2
|
ribosomal protein, large, P2
|
|
chr11_-_73309090
|
2.691
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr2_+_217277422
|
2.691
|
NM_001127207
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1
|
|
chr19_-_633563
|
2.688
|
NM_005035
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr5_+_85913755
|
2.687
|
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr11_+_57425215
|
2.681
|
NM_001142597
NM_006831
|
CLP1
|
CLP1, cleavage and polyadenylation factor I subunit, homolog (S. cerevisiae)
|
|
chr6_+_160390128
|
2.675
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr12_+_56498334
|
2.648
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr19_+_36239261
|
2.641
|
NM_019104
|
LIN37
|
lin-37 homolog (C. elegans)
|
|
chr17_-_21117899
|
2.637
|
NM_003876
|
TMEM11
|
transmembrane protein 11
|
|
chr8_-_145550536
|
2.624
|
NM_012079
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr19_-_633474
|
2.578
|
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed)
|
|
chr5_-_74807421
|
2.564
|
|
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein
|
|
chr14_+_77787705
|
2.554
|
NM_001513
|
GSTZ1
|
glutathione transferase zeta 1
|
|
chr18_+_71815745
|
2.552
|
NM_014177
|
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast)
|
|
chr1_+_40974430
|
2.536
|
NM_022774
|
DEM1
|
defects in morphology 1 homolog (S. cerevisiae)
|
|
chr1_+_151584663
|
2.535
|
|
SNX27
|
sorting nexin family member 27
|
|
chr2_+_202316391
|
2.531
|
NM_001206864
NM_018571
|
STRADB
|
STE20-related kinase adaptor beta
|
|
chr12_-_46663207
|
2.512
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr19_+_50145369
|
2.511
|
NM_021228
|
SCAF1
|
SR-related CTD-associated factor 1
|
|
chr17_-_21117479
|
2.511
|
|
TMEM11
|
transmembrane protein 11
|
|
chr9_+_130186714
|
2.505
|
|
ZNF79
|
zinc finger protein 79
|
|
chr16_+_67880784
|
2.505
|
NM_005796
|
NUTF2
|
nuclear transport factor 2
|
|
chr1_-_2457988
|
2.500
|
NM_018216
|
PANK4
|
pantothenate kinase 4
|
|
chr12_-_46662779
|
2.493
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr14_+_65453947
|
2.483
|
|
|
|
|
chr11_+_64808375
|
2.477
|
NM_013299
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr17_-_73900934
|
2.467
|
|
MRPL38
|
mitochondrial ribosomal protein L38
|
|
chr14_+_24025188
|
2.459
|
NM_001126339
NM_024328
|
THTPA
|
thiamine triphosphatase
|
|
chr1_+_54519259
|
2.444
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr10_-_94333721
|
2.435
|
|
IDE
|
insulin-degrading enzyme
|
|
chr3_+_32148000
|
2.434
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr19_+_35225124
|
2.426
|
|
ZNF181
|
zinc finger protein 181
|
|
chr12_+_56498414
|
2.422
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr16_-_58718607
|
2.405
|
NM_018231
|
SLC38A7
|
solute carrier family 38, member 7
|
|
chr1_+_1550794
|
2.403
|
NM_001170686
NM_001170687
NM_001170688
NM_080875
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr11_-_64528186
|
2.384
|
NM_001164716
NM_005609
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr18_+_7231136
|
2.380
|
NM_001105581
|
LRRC30
|
leucine rich repeat containing 30
|
|
chr12_-_64616032
|
2.375
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chrX_-_106362018
|
2.367
|
|
RBM41
|
RNA binding motif protein 41
|
|
chr18_+_77439843
|
2.365
|
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr16_+_3074060
|
2.362
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr13_-_41768533
|
2.354
|
NM_032138
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr12_+_56498324
|
2.354
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr17_+_78518742
|
2.342
|
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr9_-_116037683
|
2.326
|
|
FKBP15
|
FK506 binding protein 15, 133kDa
|
|
chr16_-_67840346
|
2.316
|
|
RANBP10
|
RAN binding protein 10
|
|
chr7_+_2394508
|
2.308
|
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr12_-_46662709
|
2.306
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr5_+_175792444
|
2.279
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr16_+_3074051
|
2.279
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr19_+_36239445
|
2.265
|
|
LIN37
|
lin-37 homolog (C. elegans)
|
|
chr19_+_58962954
|
2.261
|
NM_207395
|
ZNF324B
|
zinc finger protein 324B
|
|
chr1_-_185126153
|
2.260
|
NM_001202423
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr7_-_148823295
|
2.257
|
|
ZNF425
|
zinc finger protein 425
|
|
chr16_-_68344785
|
2.238
|
NM_032178
|
SLC7A6OS
|
solute carrier family 7, member 6 opposite strand
|
|
chr12_+_56498347
|
2.227
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr12_-_122985471
|
2.220
|
NM_017612
|
ZCCHC8
|
zinc finger, CCHC domain containing 8
|
|
chr1_-_186344383
|
2.214
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr16_-_3285359
|
2.212
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr3_+_50654868
|
2.208
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr1_-_26324534
|
2.199
|
NM_000437
|
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa
|
|
chr11_+_64808395
|
2.193
|
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr12_-_122985396
|
2.184
|
|
ZCCHC8
|
zinc finger, CCHC domain containing 8
|
|
chr2_-_69870976
|
2.182
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr19_-_2456870
|
2.181
|
|
LMNB2
|
lamin B2
|
|
chr19_+_7600581
|
2.180
|
NM_001166114
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr11_+_16760147
|
2.180
|
NM_014267
|
C11orf58
|
chromosome 11 open reading frame 58
|
|
chr7_+_2394546
|
2.179
|
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr5_-_74807805
|
2.176
|
NM_001130105
|
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein
|
|
chr1_-_243418482
|
2.172
|
|
CEP170
|
centrosomal protein 170kDa
|
|
chr12_-_57146081
|
2.171
|
NM_000946
|
PRIM1
|
primase, DNA, polypeptide 1 (49kDa)
|
|
chr4_-_88141614
|
2.170
|
NM_020803
|
KLHL8
|
kelch-like 8 (Drosophila)
|
|
chr3_+_196466806
|
2.168
|
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr7_-_135194823
|
2.153
|
NM_001008225
NM_001190847
NM_001190848
NM_001190849
NM_001190850
NM_013316
|
CNOT4
|
CCR4-NOT transcription complex, subunit 4
|
|
chr5_-_74807448
|
2.144
|
NM_005713
NM_031361
|
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein
|
|
chr17_+_78518611
|
2.142
|
NM_001163034
NM_020761
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr13_-_114144985
|
2.142
|
NM_001014283
|
DCUN1D2
|
DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae)
|
|
chr19_-_19431317
|
2.139
|
NM_172231
|
SUGP1
|
SURP and G patch domain containing 1
|
|
chr15_+_90895476
|
2.139
|
NM_001004309
|
ZNF774
|
zinc finger protein 774
|
|
chr11_-_61100444
|
2.136
|
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa
|
|
chr20_+_46130656
|
2.130
|
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr17_+_900329
|
2.125
|
NM_013337
|
TIMM22
|
translocase of inner mitochondrial membrane 22 homolog (yeast)
|
|
chr14_+_65453605
|
2.122
|
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr10_-_13390189
|
2.120
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr1_+_33938231
|
2.120
|
NM_145238
|
ZSCAN20
|
zinc finger and SCAN domain containing 20
|
|
chr7_+_2394498
|
2.116
|
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr22_+_18593556
|
2.107
|
|
TUBA8
|
tubulin, alpha 8
|
|
chr19_-_39390168
|
2.097
|
NM_012237
NM_001193286
NM_030593
|
SIRT2
|
sirtuin 2
|
|
chr7_-_75988276
|
2.096
|
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr13_-_50366703
|
2.096
|
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr10_-_94333828
|
2.095
|
|
IDE
|
insulin-degrading enzyme
|
|
chr18_-_32924417
|
2.090
|
NM_006965
|
ZNF24
|
zinc finger protein 24
|
|
chr9_-_116172572
|
2.081
|
|
POLE3
|
polymerase (DNA directed), epsilon 3 (p17 subunit)
|
|
chr2_+_103353341
|
2.078
|
|
TMEM182
|
transmembrane protein 182
|
|
chr18_+_77439798
|
2.071
|
NM_004715
NM_048368
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr1_+_84764048
|
2.068
|
NM_001010971
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr3_+_32148136
|
2.058
|
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chrX_-_106362050
|
2.056
|
NM_001171080
NM_018301
|
RBM41
|
RNA binding motif protein 41
|
|
chr16_+_3074041
|
2.054
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr13_-_41768392
|
2.054
|
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr16_-_18937722
|
2.051
|
NM_015092
|
SMG1
|
smg-1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|