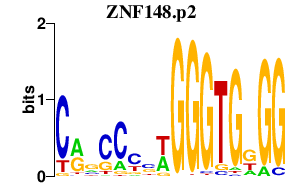
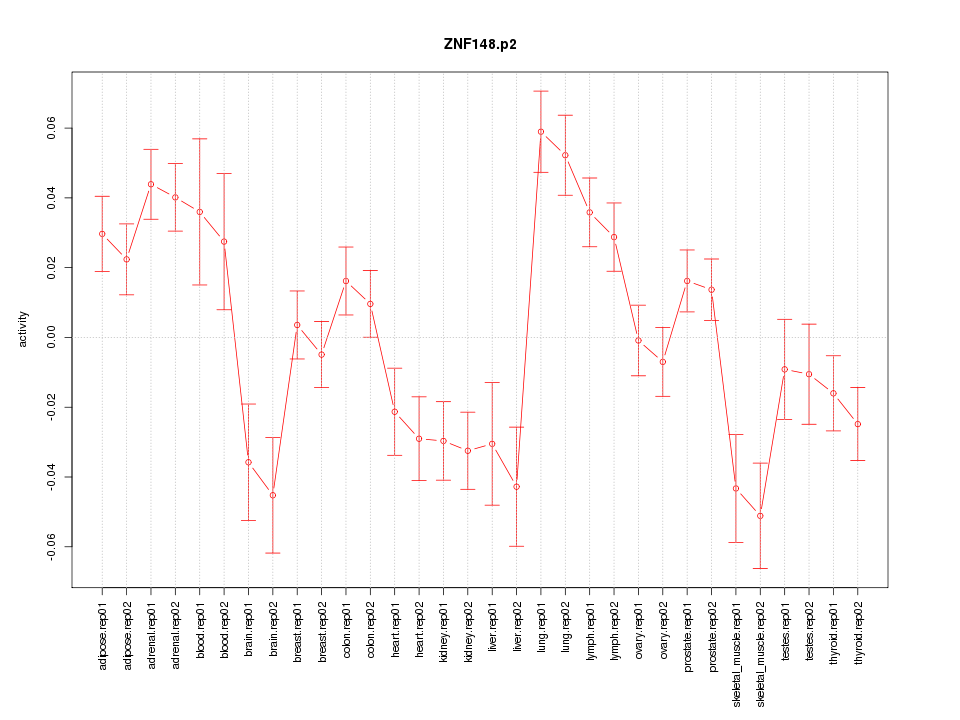
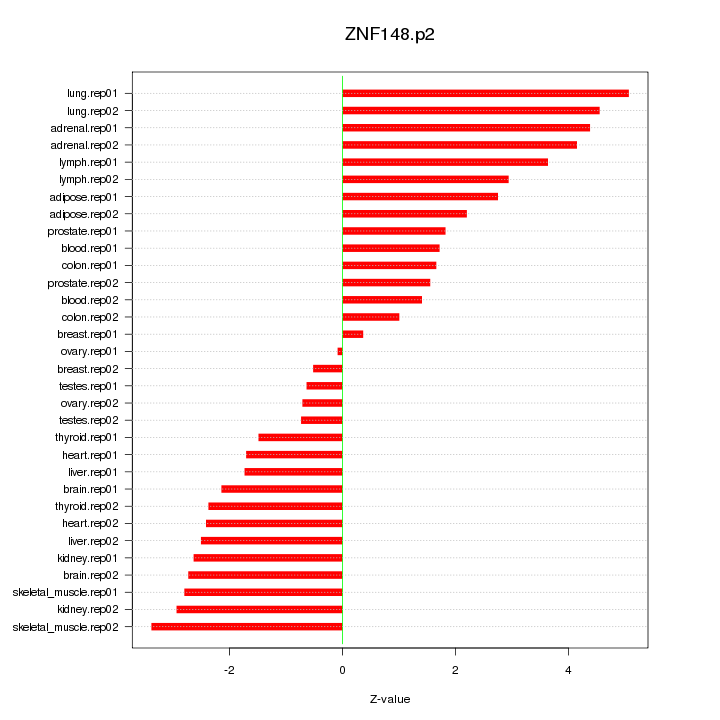
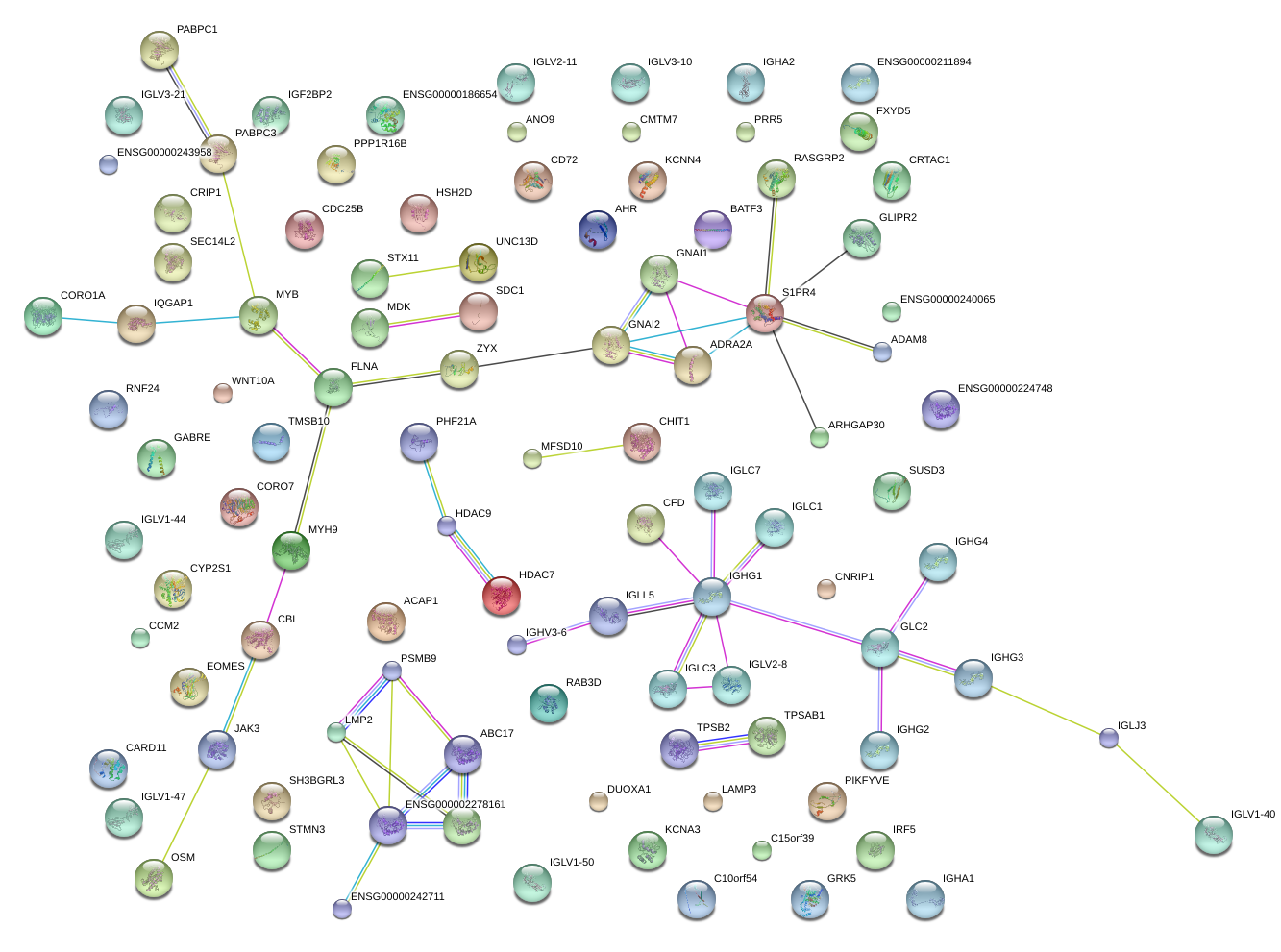
|
chr22_+_22735134
|
4.196
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr22_+_22735207
|
3.725
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr22_+_22735202
|
3.563
|
|
|
|
|
chr4_-_2936458
|
3.542
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr20_+_3776905
|
3.512
|
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr14_-_106209380
|
3.290
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_-_73533196
|
3.222
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr14_-_106092256
|
3.105
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr17_+_7239847
|
3.089
|
NM_014716
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|
|
chr22_+_23134980
|
3.004
|
|
|
|
|
chr1_-_203198772
|
2.979
|
NM_003465
|
CHIT1
|
chitinase 1 (chitotriosidase)
|
|
chr6_-_32821592
|
2.941
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr14_-_106111114
|
2.834
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr11_-_441956
|
2.828
|
|
ANO9
|
anoctamin 9
|
|
chrX_-_153599719
|
2.805
|
|
FLNA
|
filamin A, alpha
|
|
chr4_-_2936563
|
2.782
|
NM_001146069
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr10_-_135090325
|
2.772
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr11_-_441995
|
2.739
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chr16_+_30194925
|
2.727
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr22_+_22712091
|
2.666
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr16_-_4466520
|
2.654
|
|
CORO7
|
coronin 7
|
|
chrX_-_153599581
|
2.652
|
|
FLNA
|
filamin A, alpha
|
|
chr1_+_26606443
|
2.631
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr16_+_30194730
|
2.628
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr20_+_37434294
|
2.549
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr9_-_35618300
|
2.521
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr1_+_26606586
|
2.520
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr15_+_75494235
|
2.480
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr1_+_26606580
|
2.475
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr16_+_1290677
|
2.467
|
NM_003294
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr6_-_32821743
|
2.456
|
NM_000593
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr14_-_106110894
|
2.455
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr2_+_85132762
|
2.444
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr15_+_75494196
|
2.435
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr15_+_75494233
|
2.343
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr3_-_185542807
|
2.324
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr10_-_99790346
|
2.307
|
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr4_-_2935851
|
2.304
|
NM_001120
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr19_+_35645614
|
2.292
|
NM_014164
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chrX_-_153602998
|
2.292
|
NM_001110556
NM_001456
|
FLNA
|
filamin A, alpha
|
|
chr7_-_3083346
|
2.282
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr19_+_35645640
|
2.272
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr19_+_35646006
|
2.264
|
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr9_+_95821007
|
2.250
|
|
SUSD3
|
sushi domain containing 3
|
|
chr19_-_44285407
|
2.239
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr1_+_26606621
|
2.227
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr19_+_16244837
|
2.199
|
NM_032855
|
HSH2D
|
hematopoietic SH2 domain containing
|
|
chr22_-_30662787
|
2.192
|
NM_020530
|
OSM
|
oncostatin M
|
|
chrX_-_153602928
|
2.191
|
|
FLNA
|
filamin A, alpha
|
|
chr14_+_105953531
|
2.188
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr4_-_2935650
|
2.184
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr6_-_32821412
|
2.176
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr12_-_48213567
|
2.164
|
NM_001098416
NM_015401
|
HDAC7
|
histone deacetylase 7
|
|
chr16_-_4466607
|
2.161
|
|
CORO7
|
coronin 7
|
|
chr6_+_144471631
|
2.142
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr6_+_32821924
|
2.108
|
NM_002800
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2)
|
|
chr3_-_185542744
|
2.105
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr7_+_17338182
|
2.104
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr19_-_44285012
|
2.103
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr14_-_106452693
|
2.028
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr22_-_36783851
|
2.008
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr2_-_20424627
|
1.965
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr8_-_101734290
|
1.936
|
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr2_+_85132999
|
1.936
|
|
TMSB10
|
thymosin beta 10
|
|
chr22_+_45098091
|
1.930
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr10_+_120967094
|
1.922
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr20_-_3996035
|
1.914
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr19_+_41699140
|
1.911
|
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr8_-_101734313
|
1.904
|
NM_002568
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr11_+_119077021
|
1.894
|
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr3_-_182880463
|
1.890
|
NM_014398
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr14_-_106733392
|
1.887
|
|
IGHG1
IGHG3
|
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr3_+_50273745
|
1.875
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr1_-_111217625
|
1.871
|
NM_002232
|
KCNA3
|
potassium voltage-gated channel, shaker-related subfamily, member 3
|
|
chr1_-_212873104
|
1.869
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr7_+_128577990
|
1.859
|
NM_001098629
NM_001098630
NM_001242452
|
IRF5
|
interferon regulatory factor 5
|
|
chr19_-_11450254
|
1.858
|
NM_004283
|
RAB3D
|
RAB3D, member RAS oncogene family
|
|
chr22_+_45098046
|
1.855
|
NM_181333
|
PRR5
|
proline rich 5 (renal)
|
|
chr22_-_36783977
|
1.853
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr14_-_106174914
|
1.842
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr22_-_36783930
|
1.822
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr15_-_45422056
|
1.813
|
NM_144565
|
DUOXA1
|
dual oxidase maturation factor 1
|
|
chr3_-_27764197
|
1.812
|
|
EOMES
|
eomesodermin
|
|
chr22_+_23165273
|
1.806
|
|
|
|
|
chr11_+_46402333
|
1.799
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr11_-_64511550
|
1.796
|
NM_001098671
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr17_-_73840422
|
1.795
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr10_+_112836784
|
1.788
|
NM_000681
|
ADRA2A
|
adrenergic, alpha-2A-, receptor
|
|
chr15_+_90931449
|
1.766
|
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr20_-_62284701
|
1.764
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr19_+_3178735
|
1.761
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr11_-_46142955
|
1.744
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr15_+_90931470
|
1.742
|
NM_003870
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr9_+_36136710
|
1.740
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chrX_-_151143096
|
1.734
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chr6_+_135502521
|
1.734
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr1_-_161039745
|
1.730
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr19_+_859658
|
1.727
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr3_+_32433342
|
1.726
|
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7
|
|
chr19_+_859673
|
1.715
|
|
CFD
|
complement factor D (adipsin)
|
|
chr6_+_135502445
|
1.702
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr19_-_17958814
|
1.700
|
NM_000215
|
JAK3
|
Janus kinase 3
|
|
chr2_+_219745243
|
1.696
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr7_+_143058794
|
1.685
|
|
ZYX
|
zyxin
|
|
chr6_-_31704070
|
1.678
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr3_-_133748828
|
1.656
|
NM_005630
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr18_+_12308236
|
1.655
|
|
TUBB6
|
tubulin, beta 6 class V
|
|
chr19_-_14247267
|
1.655
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr1_-_161039649
|
1.639
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr5_-_171615095
|
1.637
|
|
STK10
|
serine/threonine kinase 10
|
|
chr3_+_32433151
|
1.635
|
NM_138410
NM_181472
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7
|
|
chr19_+_54371125
|
1.635
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr9_-_123691442
|
1.621
|
NM_001190945
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr1_+_26856240
|
1.615
|
NM_002953
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr7_+_98972326
|
1.607
|
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa
|
|
chr1_-_161039413
|
1.605
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr19_-_41859830
|
1.602
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr20_-_4804067
|
1.602
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr22_+_25960738
|
1.584
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr22_-_37882214
|
1.584
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr12_-_122231554
|
1.572
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chr20_-_3748355
|
1.571
|
NM_001039140
|
C20orf27
|
chromosome 20 open reading frame 27
|
|
chr4_-_7941092
|
1.569
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr22_+_44577215
|
1.568
|
|
PARVG
|
parvin, gamma
|
|
chr6_+_35265594
|
1.564
|
NM_022047
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr1_-_161039703
|
1.562
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr14_-_60337350
|
1.560
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chrX_-_48327739
|
1.557
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr1_-_212873306
|
1.556
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr1_-_6321034
|
1.554
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr7_+_98972351
|
1.551
|
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa
|
|
chr6_-_31704093
|
1.549
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr7_+_76139744
|
1.547
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr19_+_14142572
|
1.544
|
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr1_-_212873197
|
1.542
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr17_-_76124791
|
1.535
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr19_+_14142611
|
1.520
|
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr9_+_95820948
|
1.516
|
NM_145006
|
SUSD3
|
sushi domain containing 3
|
|
chr22_+_17565920
|
1.515
|
|
IL17RA
|
interleukin 17 receptor A
|
|
chr19_-_41859767
|
1.511
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr9_-_136344182
|
1.510
|
NM_001145099
NM_017585
|
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6
|
|
chr9_-_140064426
|
1.507
|
NM_001013653
|
LRRC26
|
leucine rich repeat containing 26
|
|
chr19_-_14247400
|
1.507
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr11_-_2323066
|
1.500
|
NM_001142946
|
C11orf21
|
chromosome 11 open reading frame 21
|
|
chr22_-_50970542
|
1.496
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr16_-_1280162
|
1.485
|
NM_024164
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr19_+_54371150
|
1.482
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr15_+_65134081
|
1.475
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr15_+_90931489
|
1.474
|
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr3_+_108541544
|
1.454
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr17_-_73840587
|
1.454
|
NM_199242
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr15_+_90931505
|
1.454
|
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr19_+_48828820
|
1.454
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr19_+_54371170
|
1.452
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_+_19971865
|
1.442
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr3_+_47021146
|
1.441
|
NM_015175
|
NBEAL2
|
neurobeachin-like 2
|
|
chr22_+_39853316
|
1.437
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chrX_+_129305553
|
1.423
|
NM_004794
|
RAB33A
|
RAB33A, member RAS oncogene family
|
|
chr17_+_77751929
|
1.422
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr8_-_101734162
|
1.421
|
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr2_+_192542860
|
1.415
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr22_-_36784055
|
1.411
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr5_+_141016567
|
1.411
|
|
RELL2
|
RELT-like 2
|
|
chr1_-_89738477
|
1.404
|
NM_001134486
NM_052942
|
GBP5
|
guanylate binding protein 5
|
|
chr1_+_19972259
|
1.400
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr15_-_34629941
|
1.398
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr9_-_130517552
|
1.397
|
NM_001142534
NM_001142533
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr19_+_16187266
|
1.397
|
|
TPM4
|
tropomyosin 4
|
|
chr1_-_38397417
|
1.390
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr19_+_6531009
|
1.389
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr15_+_90931509
|
1.386
|
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr15_-_34630203
|
1.386
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr19_+_41116069
|
1.386
|
|
|
|
|
chr17_-_47308108
|
1.383
|
NM_001143804
NM_178500
|
PHOSPHO1
|
phosphatase, orphan 1
|
|
chr5_+_40680021
|
1.378
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr19_+_49199227
|
1.377
|
NM_000511
NM_001097638
|
FUT2
|
fucosyltransferase 2 (secretor status included)
|
|
chr22_+_44577256
|
1.374
|
NM_001137606
|
PARVG
|
parvin, gamma
|
|
chr11_-_68780667
|
1.371
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr22_-_36784001
|
1.370
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr3_-_133748727
|
1.368
|
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr11_-_5271035
|
1.364
|
NM_000559
|
HBG1
|
hemoglobin, gamma A
|
|
chr3_-_182880432
|
1.363
|
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr11_+_64107687
|
1.361
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr16_-_4466595
|
1.360
|
|
CORO7
|
coronin 7
|
|
chr11_-_68518944
|
1.358
|
NM_001039656
NM_004923
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin)
|
|
chr6_+_30457182
|
1.347
|
NM_005516
|
HLA-E
|
major histocompatibility complex, class I, E
|
|
chr19_+_14142459
|
1.346
|
|
IL27RA
|
interleukin 27 receptor, alpha
|
|
chr10_+_112257624
|
1.345
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr5_+_156887204
|
1.344
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr11_-_65625650
|
1.342
|
|
|
|
|
chr7_+_77166750
|
1.341
|
NM_002835
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr20_-_62711160
|
1.340
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr9_-_100954883
|
1.339
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr22_+_25960885
|
1.338
|
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr16_-_67427335
|
1.338
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr1_-_161039586
|
1.335
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr7_+_143079925
|
1.326
|
|
ZYX
|
zyxin
|
|
chr9_+_36036900
|
1.325
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr17_+_74380676
|
1.324
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr3_+_43328001
|
1.322
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|