|
chr4_+_74735107
|
2.071
|
NM_001511
|
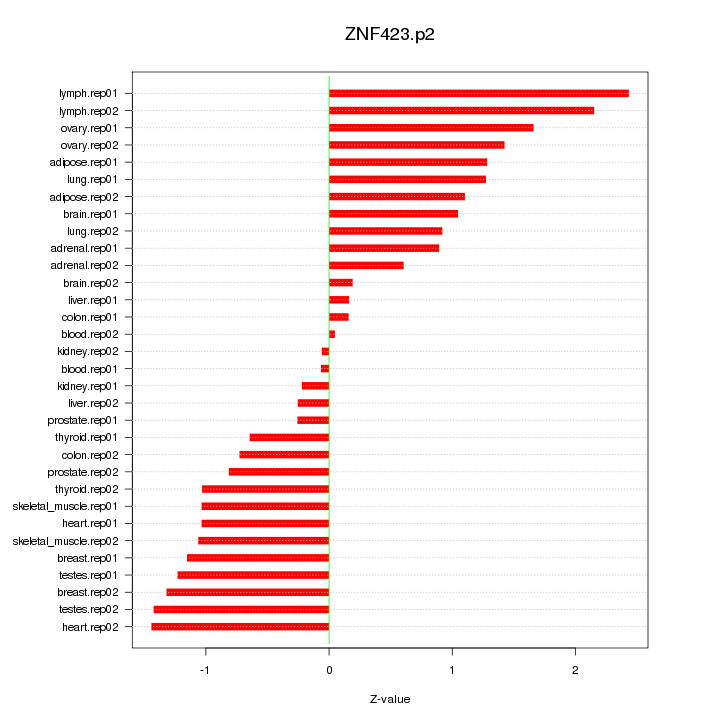
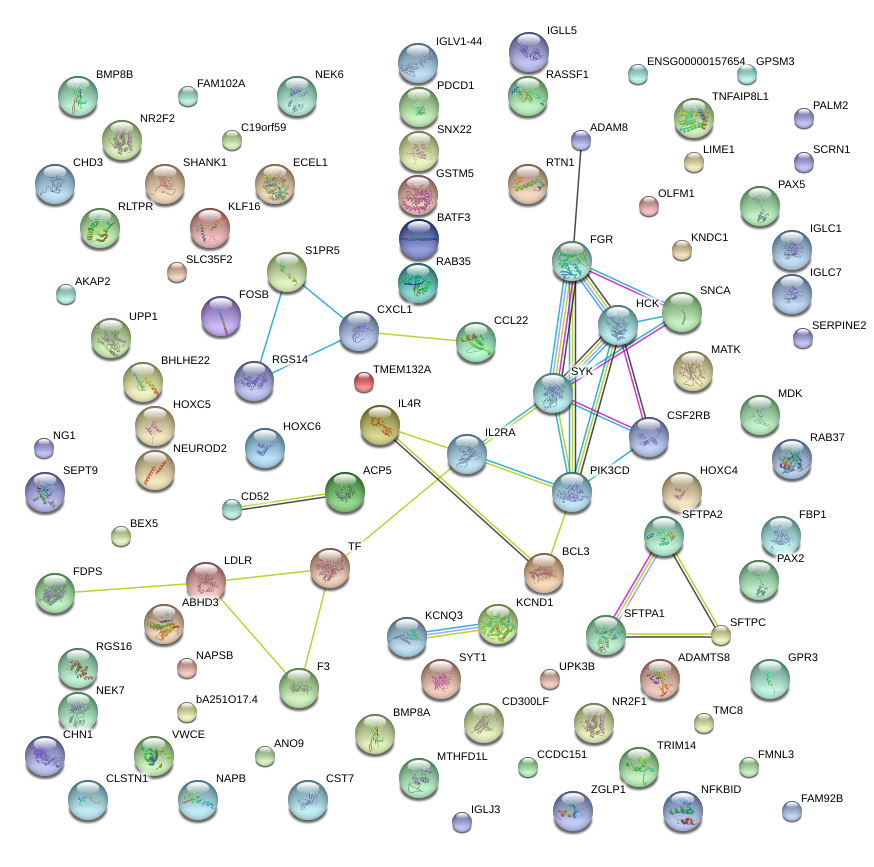
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr19_-_11689765
|
1.351
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr10_+_81370694
|
1.309
|
NM_001093770
NM_001164644
NM_001164645
NM_001164646
NM_001164647
NM_005411
|
SFTPA1
|
surfactant protein A1
|
|
chr19_-_36391448
|
1.304
|
NM_139239
|
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta
|
|
chrX_-_48828250
|
1.263
|
NM_004979
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1
|
|
chr11_+_60691911
|
1.257
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr16_-_85145995
|
1.233
|
NM_198491
|
FAM92B
|
family with sequence similarity 92, member B
|
|
chr5_+_92920592
|
1.193
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr5_+_176784843
|
1.132
|
NM_006480
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr8_+_65493610
|
1.091
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr1_+_26644458
|
1.083
|
|
CD52
|
CD52 molecule
|
|
chr19_-_3786279
|
1.058
|
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr10_-_135090325
|
1.037
|
NM_001109
NM_001164489
NM_001164490
|
ADAM8
|
ADAM metallopeptidase domain 8
|
|
chr1_-_27952591
|
1.032
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr22_+_22786283
|
1.031
|
|
|
|
|
chr14_-_106714594
|
1.007
|
|
|
|
|
chr19_-_11689591
|
0.993
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr1_+_27719151
|
0.992
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr19_+_11200146
|
0.969
|
|
LDLR
|
low density lipoprotein receptor
|
|
chr19_+_11200124
|
0.952
|
|
LDLR
|
low density lipoprotein receptor
|
|
chr6_+_151186884
|
0.927
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr8_+_22019167
|
0.874
|
NM_001172357
NM_001172410
NM_003018
|
SFTPC
|
surfactant protein C
|
|
chr17_+_72733350
|
0.855
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr2_-_224903933
|
0.847
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr17_-_72709010
|
0.841
|
NM_139018
|
CD300LF
|
CD300 molecule-like family member f
|
|
chr1_-_212873197
|
0.833
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr20_+_24929862
|
0.827
|
NM_003650
|
CST7
|
cystatin F (leukocystatin)
|
|
chr21_+_26934440
|
0.811
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chrX_-_101410761
|
0.778
|
NM_001159560
NM_001012978
|
BEX5
|
brain expressed, X-linked 5
|
|
chr16_+_67679011
|
0.774
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr19_-_10420220
|
0.774
|
NM_001103167
|
ZGLP1
|
zinc finger, GATA-like protein 1
|
|
chr19_+_45971249
|
0.773
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr19_+_14247974
|
0.768
|
|
LOC100507373
|
uncharacterized LOC100507373
|
|
chr1_+_26644380
|
0.765
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr7_+_76139744
|
0.763
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr14_-_60097314
|
0.762
|
|
RTN1
|
reticulon 1
|
|
chr20_-_23402038
|
0.744
|
NM_022080
|
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta
|
|
chr19_+_46144904
|
0.742
|
NM_001242348
|
LOC100287177
|
uncharacterized LOC100287177
|
|
chr22_+_22735202
|
0.726
|
|
|
|
|
chr9_-_100881462
|
0.715
|
NM_014788
NM_033219
NM_033220
|
TRIM14
|
tripartite motif containing 14
|
|
chr19_+_11200037
|
0.709
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr1_+_110254863
|
0.708
|
NM_000851
|
GSTM5
|
glutathione S-transferase mu 5
|
|
chr6_+_151187064
|
0.701
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr6_+_151186814
|
0.701
|
NM_001242767
NM_001242769
NM_015440
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr19_-_11545900
|
0.697
|
NM_145045
|
CCDC151
|
coiled-coil domain containing 151
|
|
chr18_-_19284622
|
0.681
|
|
ABHD3
|
abhydrolase domain containing 3
|
|
chr17_+_76126835
|
0.679
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr9_+_93564204
|
0.676
|
NM_001174168
|
SYK
|
spleen tyrosine kinase
|
|
chr12_-_120554453
|
0.671
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr10_-_6104113
|
0.669
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr22_+_22735134
|
0.668
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr8_-_125486808
|
0.667
|
|
|
|
|
chr16_+_57392694
|
0.653
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr19_-_1863546
|
0.652
|
NM_031918
|
KLF16
|
Kruppel-like factor 16
|
|
chr19_+_45251933
|
0.642
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr19_+_4640028
|
0.640
|
NM_001167942
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1
|
|
chr9_-_130742803
|
0.640
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr5_+_176784947
|
0.638
|
|
|
|
|
chr17_+_72732958
|
0.635
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr18_-_19284737
|
0.630
|
NM_138340
|
ABHD3
|
abhydrolase domain containing 3
|
|
chr8_-_133493003
|
0.625
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr9_+_93564063
|
0.621
|
|
SYK
|
spleen tyrosine kinase
|
|
chr1_-_27953046
|
0.612
|
NM_001042729
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr9_-_37034358
|
0.610
|
NM_016734
|
PAX5
|
paired box 5
|
|
chr3_-_50374749
|
0.610
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr11_-_130298305
|
0.609
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr2_-_233352530
|
0.609
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr20_+_62368035
|
0.608
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr20_+_30639981
|
0.607
|
NM_001172129
NM_001172130
NM_001172131
NM_001172132
NM_001172133
NM_002110
|
HCK
|
hemopoietic cell kinase
|
|
chr15_-_95870310
|
0.607
|
|
LOC400456
|
uncharacterized LOC400456
|
|
chr11_-_441995
|
0.604
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chr11_+_46403279
|
0.601
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr3_-_50374795
|
0.601
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr19_-_10628111
|
0.600
|
NM_030760
|
S1PR5
|
sphingosine-1-phosphate receptor 5
|
|
chr6_-_32160289
|
0.600
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr7_+_27135983
|
0.599
|
|
|
|
|
chr19_-_3786382
|
0.599
|
NM_139354
NM_139355
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr9_+_137967401
|
0.596
|
|
OLFM1
|
olfactomedin 1
|
|
chr16_+_27325244
|
0.595
|
NM_000418
NM_001008699
|
IL4R
|
interleukin 4 receptor
|
|
chr12_-_120554551
|
0.595
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr17_-_37764165
|
0.595
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr12_-_50100966
|
0.587
|
|
|
|
|
chr11_-_441956
|
0.586
|
|
ANO9
|
anoctamin 9
|
|
chr12_-_50101127
|
0.586
|
|
FMNL3
|
formin-like 3
|
|
chr2_-_242800989
|
0.580
|
NM_005018
|
PDCD1
|
programmed cell death 1
|
|
chr7_-_30029284
|
0.579
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr7_+_48128225
|
0.576
|
NM_181597
|
UPP1
|
uridine phosphorylase 1
|
|
chr22_+_37309618
|
0.572
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr11_-_107729523
|
0.567
|
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr12_+_79258588
|
0.564
|
|
SYT1
|
synaptotagmin I
|
|
chr17_+_7788094
|
0.564
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr3_-_50374875
|
0.563
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr9_+_127019782
|
0.561
|
NM_001166167
NM_001145001
NM_014397
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr15_+_64443909
|
0.561
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr11_+_46403302
|
0.559
|
NM_001012333
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr2_-_175869910
|
0.557
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr9_+_93564099
|
0.556
|
|
SYK
|
spleen tyrosine kinase
|
|
chr9_+_93564011
|
0.556
|
NM_001135052
NM_003177
|
SYK
|
spleen tyrosine kinase
|
|
chr4_-_90758126
|
0.555
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr19_+_7741934
|
0.545
|
NM_174918
|
C19orf59
|
chromosome 19 open reading frame 59
|
|
chr20_+_24929904
|
0.543
|
|
CST7
|
cystatin F (leukocystatin)
|
|
chr1_-_182573393
|
0.536
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr20_+_62367945
|
0.535
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr1_+_155278677
|
0.532
|
NM_001135821
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr9_-_97401588
|
0.531
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr22_+_22735207
|
0.530
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr1_+_9711781
|
0.526
|
NM_005026
|
PIK3CD
|
phosphoinositide-3-kinase, catalytic, delta polypeptide
|
|
chr8_-_133492556
|
0.525
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr10_+_134973935
|
0.520
|
NM_152643
|
KNDC1
|
kinase non-catalytic C-lobe domain (KIND) containing 1
|
|
chr11_-_61062697
|
0.519
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr9_+_112542496
|
0.518
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr1_-_40254131
|
0.511
|
|
BMP8B
|
bone morphogenetic protein 8b
|
|
chr12_+_54410893
|
0.510
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr19_-_51220194
|
0.499
|
NM_016148
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1
|
|
chr12_+_54410641
|
0.498
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr3_+_133465204
|
0.496
|
|
TF
|
transferrin
|
|
chr1_-_212873306
|
0.495
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr7_+_100081549
|
0.495
|
NM_173564
|
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1
|
|
chr6_+_42897184
|
0.494
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr7_-_100171269
|
0.483
|
NM_001168682
|
SAP25
|
Sin3A-associated protein, 25kDa
|
|
chr19_+_55085237
|
0.481
|
NM_001130917
NM_006866
|
LILRA2
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2
|
|
chr2_-_145274916
|
0.481
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr7_+_26191732
|
0.479
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chr1_+_109642814
|
0.478
|
|
SCARNA2
|
small Cajal body-specific RNA 2
|
|
chr6_-_2840683
|
0.478
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr1_+_155278727
|
0.473
|
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr12_-_125348346
|
0.473
|
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr1_+_1115076
|
0.471
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr11_-_107729492
|
0.468
|
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr19_-_46476556
|
0.466
|
NM_002516
|
NOVA2
|
neuro-oncological ventral antigen 2
|
|
chr14_-_60097531
|
0.463
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr16_+_71392615
|
0.461
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr18_-_47018777
|
0.460
|
|
RPL17
|
ribosomal protein L17
|
|
chr7_-_5570188
|
0.458
|
NM_001101
|
ACTB
|
actin, beta
|
|
chr17_+_48638339
|
0.454
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr12_-_120554557
|
0.453
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr12_+_79258517
|
0.451
|
|
SYT1
|
synaptotagmin I
|
|
chr4_-_74964836
|
0.451
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr5_-_180018465
|
0.451
|
NM_052863
|
SCGB3A1
|
secretoglobin, family 3A, member 1
|
|
chr12_-_125348422
|
0.449
|
NM_001082959
NM_005505
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr22_+_35777051
|
0.448
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr19_+_38810461
|
0.448
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr17_+_7311501
|
0.447
|
NM_020795
|
NLGN2
|
neuroligin 2
|
|
chr16_+_27325269
|
0.445
|
|
IL4R
|
interleukin 4 receptor
|
|
chr9_-_100881515
|
0.445
|
|
TRIM14
|
tripartite motif containing 14
|
|
chr7_-_97601654
|
0.445
|
|
|
|
|
chr14_-_106209380
|
0.444
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr18_-_5543967
|
0.442
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr2_+_98330030
|
0.441
|
NM_001079
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chr6_-_30654978
|
0.439
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr6_-_166401526
|
0.438
|
|
LINC00473
|
long intergenic non-protein coding RNA 473
|
|
chr19_+_38810751
|
0.436
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr21_+_45719953
|
0.434
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr7_+_150102376
|
0.433
|
|
|
|
|
chr19_+_7828034
|
0.431
|
NM_001144904
NM_001144905
NM_001144906
NM_001144907
NM_001144908
NM_001144909
NM_001144910
NM_001144911
NM_014257
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr11_-_44972227
|
0.431
|
NM_001076787
NM_006034
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr7_+_50344255
|
0.431
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr12_-_6665220
|
0.430
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr6_+_42897236
|
0.430
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr19_-_41858948
|
0.429
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr3_+_6902797
|
0.429
|
NM_000844
NM_181874
|
GRM7
|
glutamate receptor, metabotropic 7
|
|
chr22_+_39853316
|
0.428
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr12_+_79258448
|
0.426
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr9_+_93564073
|
0.425
|
|
SYK
|
spleen tyrosine kinase
|
|
chr6_+_151187310
|
0.425
|
NM_001242768
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr3_-_158450205
|
0.425
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr11_+_43665772
|
0.425
|
|
|
|
|
chr19_-_10679620
|
0.424
|
NM_001800
NM_079421
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr12_-_120554558
|
0.423
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr22_+_23154240
|
0.422
|
|
|
|
|
chr20_+_57766074
|
0.422
|
NM_178457
|
ZNF831
|
zinc finger protein 831
|
|
chr12_-_50297575
|
0.419
|
NM_012306
|
FAIM2
|
Fas apoptotic inhibitory molecule 2
|
|
chr12_-_125348266
|
0.411
|
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr6_+_31543340
|
0.410
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr1_-_85462622
|
0.409
|
NM_153259
|
MCOLN2
|
mucolipin 2
|
|
chr10_+_102792161
|
0.407
|
|
SFXN3
|
sideroflexin 3
|
|
chr19_+_18530423
|
0.405
|
|
|
|
|
chr19_-_1863425
|
0.400
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr6_+_42896859
|
0.399
|
NM_006586
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr19_+_38810540
|
0.397
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr17_+_39969205
|
0.396
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr14_-_60097223
|
0.395
|
|
RTN1
|
reticulon 1
|
|
chr17_+_7608413
|
0.393
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chrX_-_153881757
|
0.393
|
NM_020994
NM_172377
|
CTAG2
|
cancer/testis antigen 2
|
|
chr19_-_48673559
|
0.392
|
NM_000234
|
LIG1
|
ligase I, DNA, ATP-dependent
|
|
chr12_+_65218351
|
0.387
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr1_-_102462789
|
0.386
|
NM_058170
|
OLFM3
|
olfactomedin 3
|
|
chr19_-_36523540
|
0.386
|
NM_001199570
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr14_-_106518424
|
0.385
|
|
IGH@
IGHA1
IGHG1
IGHG2
IGHM
|
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant mu
|
|
chr4_-_46391771
|
0.385
|
NM_000807
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr2_+_74776146
|
0.385
|
NM_001197260
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr10_+_102792166
|
0.385
|
|
SFXN3
|
sideroflexin 3
|
|
chr2_-_198175324
|
0.376
|
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr1_+_210406194
|
0.374
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr1_+_67218148
|
0.374
|
|
TCTEX1D1
|
Tctex1 domain containing 1
|
|
chr19_+_7733971
|
0.372
|
NM_001193374
NM_020415
|
RETN
|
resistin
|
|
chr11_-_47400096
|
0.372
|
NM_001080547
NM_003120
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr4_+_4387982
|
0.371
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr22_+_35777087
|
0.371
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr20_+_44650260
|
0.368
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|