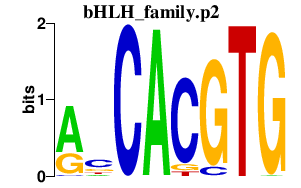
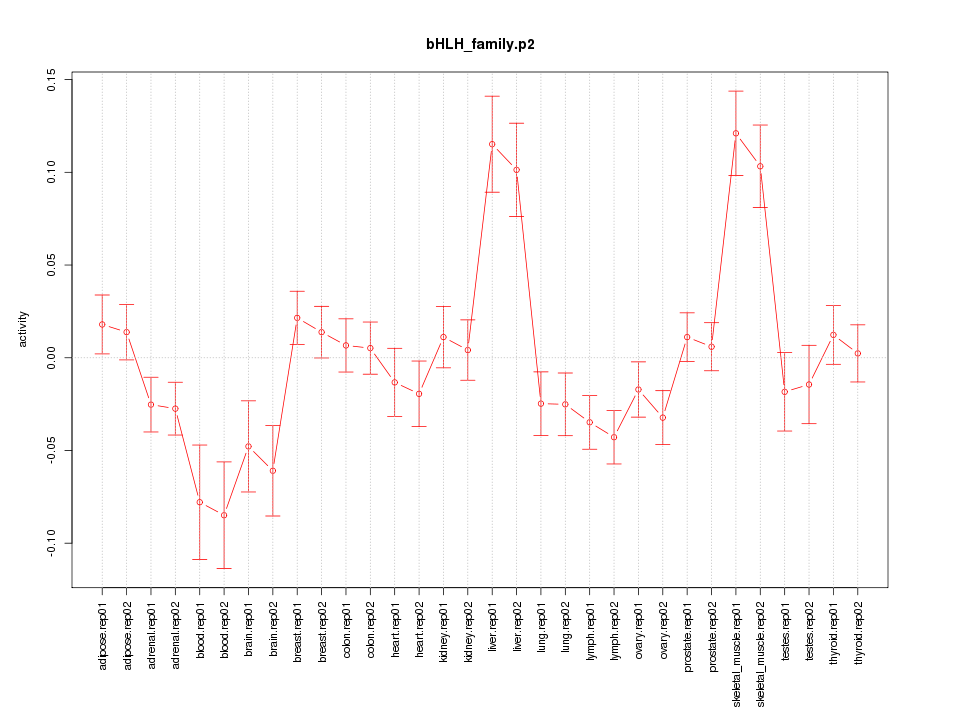
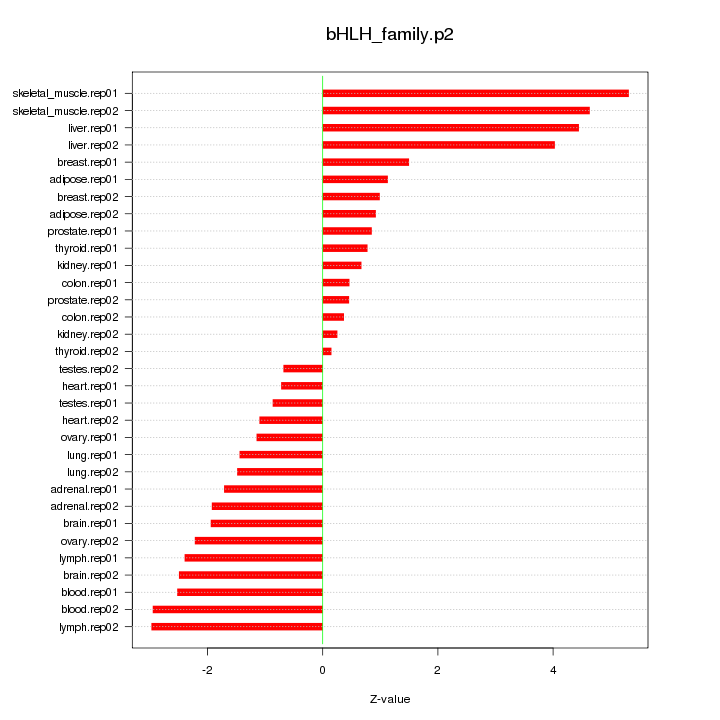
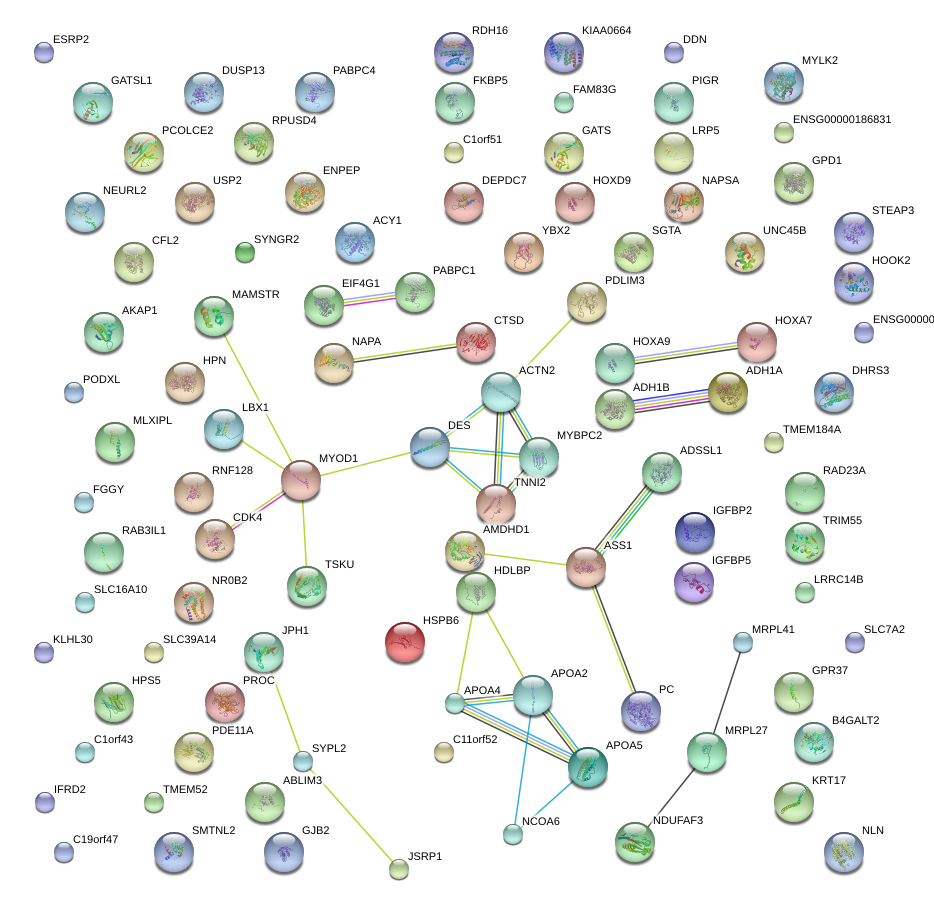
|
chr12_+_50497790
|
10.789
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr8_-_75233461
|
9.657
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr11_-_116663106
|
7.429
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr1_-_1850704
|
7.365
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr9_+_133320093
|
6.814
|
NM_000050
NM_054012
|
ASS1
|
argininosuccinate synthase 1
|
|
chr4_-_100242487
|
5.385
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr9_+_133320247
|
5.375
|
|
ASS1
|
argininosuccinate synthase 1
|
|
chr1_-_161193375
|
5.202
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr11_-_2018568
|
5.186
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_-_2018703
|
5.010
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_-_154193073
|
4.862
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr1_+_150255228
|
4.747
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr11_-_2018446
|
4.712
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr20_+_30407175
|
4.645
|
NM_033118
|
MYLK2
|
myosin light chain kinase 2
|
|
chr11_-_119252364
|
4.565
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_154192882
|
4.450
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr8_+_17354562
|
4.393
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr11_+_111789600
|
4.370
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr11_-_66725790
|
4.272
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr1_+_110009099
|
4.236
|
NM_001040709
|
SYPL2
|
synaptophysin-like 2
|
|
chr11_-_119252267
|
4.174
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr11_-_126081532
|
4.132
|
NM_001144827
NM_032795
|
RPUSD4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr8_+_67039130
|
4.084
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr11_+_76494132
|
4.023
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr1_-_27240430
|
3.903
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr19_-_36246411
|
3.844
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr12_-_58146110
|
3.843
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr19_+_35532617
|
3.693
|
|
HPN
|
hepsin
|
|
chr17_-_7197866
|
3.691
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr11_+_68079999
|
3.682
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr11_-_2018334
|
3.681
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr19_-_36246206
|
3.659
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr6_-_35696359
|
3.602
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr5_+_191625
|
3.598
|
NM_001080478
|
LRRC14B
|
leucine rich repeat containing 14B
|
|
chr12_-_58146025
|
3.560
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_+_110009220
|
3.550
|
|
SYPL2
|
synaptophysin-like 2
|
|
chr17_+_76164624
|
3.475
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr17_-_7197811
|
3.455
|
|
YBX2
|
Y box binding protein 2
|
|
chr11_+_111789658
|
3.452
|
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr12_-_57351381
|
3.435
|
NM_003708
|
RDH16
|
retinol dehydrogenase 16 (all-trans)
|
|
chr11_-_18343670
|
3.407
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr19_-_40854283
|
3.393
|
NM_178830
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr12_-_49392909
|
3.336
|
NM_015086
|
DDN
|
dendrin
|
|
chr19_-_49222956
|
3.279
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr1_-_154193044
|
3.255
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr12_-_58146108
|
3.240
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr14_-_35183722
|
3.222
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr11_-_61684981
|
3.219
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr5_+_65017993
|
3.218
|
NM_020726
|
NLN
|
neurolysin (metallopeptidase M3 family)
|
|
chr17_+_4487833
|
3.211
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr17_-_2614926
|
3.174
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr11_+_33037727
|
3.173
|
NM_139160
|
DEPDC7
|
DEP domain containing 7
|
|
chr7_-_27196163
|
3.149
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr8_-_17658711
|
3.132
|
|
|
|
|
chr3_+_52017299
|
3.087
|
NM_000666
NM_001198895
NM_001198896
NM_001198897
NM_001198898
|
ACY1
|
aminoacylase 1
|
|
chr19_-_40854410
|
3.086
|
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr19_+_50936159
|
3.071
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr2_-_217559497
|
3.057
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr17_+_55163077
|
2.920
|
NM_001242903
NM_001242902
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr6_+_111408780
|
2.912
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr19_-_2256415
|
2.904
|
NM_144616
|
JSRP1
|
junctional sarcoplasmic reticulum protein 1
|
|
chr2_+_119981389
|
2.901
|
|
STEAP3
|
STEAP family member 3, metalloreductase
|
|
chr13_-_20767036
|
2.857
|
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr7_-_27205075
|
2.851
|
|
HOXA9
|
homeobox A9
|
|
chr3_+_49059046
|
2.828
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr3_+_49059490
|
2.808
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr2_+_119981383
|
2.807
|
NM_001008410
NM_018234
NM_182915
|
STEAP3
|
STEAP family member 3, metalloreductase
|
|
chr3_-_142607737
|
2.777
|
NM_013363
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2
|
|
chr2_-_178937472
|
2.775
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr22_-_41593466
|
2.745
|
|
|
|
|
chr7_-_1595746
|
2.741
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr11_+_1860232
|
2.738
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr8_+_22224997
|
2.738
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr17_+_33474835
|
2.733
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr3_-_50329702
|
2.730
|
NM_006764
|
IFRD2
|
interferon-related developmental regulator 2
|
|
chr17_-_18907988
|
2.729
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr1_-_207119707
|
2.718
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr19_+_13056656
|
2.716
|
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr20_-_44519629
|
2.707
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr1_+_44445548
|
2.700
|
NM_030587
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr3_+_184032319
|
2.685
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr1_-_161193190
|
2.663
|
|
APOA2
|
apolipoprotein A-II
|
|
chr3_+_184032243
|
2.658
|
NM_001194946
NM_001194947
NM_198241
NM_198242
NM_198244
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr1_-_12677347
|
2.655
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr4_+_111397181
|
2.651
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr12_+_96337070
|
2.630
|
NM_152435
|
AMDHD1
|
amidohydrolase domain containing 1
|
|
chr2_+_217498082
|
2.617
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr10_-_76868853
|
2.616
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr2_+_220283098
|
2.612
|
NM_001927
|
DES
|
desmin
|
|
chr8_+_22224758
|
2.605
|
NM_001128431
NM_001135154
NM_015359
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr1_+_59762486
|
2.597
|
NM_001113411
NM_018291
|
FGGY
|
FGGY carbohydrate kinase domain containing
|
|
chr10_-_102988688
|
2.591
|
NM_006562
|
LBX1
|
ladybird homeobox 1
|
|
chr3_+_184032414
|
2.589
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr14_+_105190533
|
2.588
|
NM_152328
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr2_+_239047362
|
2.588
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr7_-_73038814
|
2.571
|
NM_032951
NM_032952
NM_032953
NM_032954
|
MLXIPL
|
MLX interacting protein-like
|
|
chr4_-_186456580
|
2.549
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr2_+_172864803
|
2.543
|
NM_199227
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial)
|
|
chr16_-_68269948
|
2.537
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr17_-_48450532
|
2.528
|
NM_016504
|
MRPL27
|
mitochondrial ribosomal protein L27
|
|
chr11_-_116693907
|
2.528
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr1_+_236849753
|
2.523
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr8_+_22225173
|
2.522
|
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr12_-_58146118
|
2.503
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr11_+_17741109
|
2.499
|
NM_002478
|
MYOD1
|
myogenic differentiation 1
|
|
chr5_+_148520985
|
2.497
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr7_+_74379079
|
2.497
|
NM_001145063
|
GATSL1
GATSL2
|
GATS protein-like 1
GATS protein-like 2
|
|
chr19_-_2783332
|
2.491
|
NM_003021
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr19_-_48018317
|
2.485
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr19_-_12886263
|
2.476
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr11_-_2017822
|
2.459
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr7_-_124405029
|
2.437
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chrX_+_105937067
|
2.433
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr11_-_1785126
|
2.425
|
|
CTSD
|
cathepsin D
|
|
chr1_+_182992694
|
2.424
|
|
|
|
|
chr2_-_242255114
|
2.405
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr1_-_40042072
|
2.396
|
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr12_+_6833425
|
2.385
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr22_+_35695872
|
2.384
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr1_+_26496382
|
2.374
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr11_-_64014381
|
2.363
|
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chr8_+_120220554
|
2.349
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr16_+_71660063
|
2.348
|
NM_001017967
NM_052858
|
MARVELD3
|
MARVEL domain containing 3
|
|
chr6_-_33714681
|
2.344
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr2_-_242254984
|
2.334
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr1_+_44444986
|
2.321
|
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr7_-_73184535
|
2.321
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr11_-_64014264
|
2.308
|
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chr2_-_152590983
|
2.295
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr10_-_82049178
|
2.288
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr19_-_48018163
|
2.279
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr2_-_208634051
|
2.275
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr6_-_35656677
|
2.258
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr22_+_40390935
|
2.250
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr14_-_24657986
|
2.244
|
|
IPO4
|
importin 4
|
|
chr1_-_159915382
|
2.242
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr12_+_54393876
|
2.240
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr10_-_75410778
|
2.222
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr7_-_111202346
|
2.221
|
NM_032549
NM_001244606
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae)
|
|
chr10_-_93392857
|
2.220
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr10_-_93392799
|
2.218
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr3_+_184032849
|
2.217
|
NM_182917
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr1_+_167905796
|
2.193
|
NM_001017977
NM_001198956
NM_001198957
NM_018442
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr10_-_93392744
|
2.183
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr1_+_9599546
|
2.157
|
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr11_-_63933493
|
2.151
|
NM_014067
|
MACROD1
|
MACRO domain containing 1
|
|
chr5_-_150466696
|
2.148
|
NM_001252391
NM_001252392
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr1_-_145075636
|
2.133
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_151804225
|
2.123
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr22_+_35695792
|
2.122
|
NM_001135730
NM_001135732
NM_005488
|
TOM1
|
target of myb1 (chicken)
|
|
chr19_+_35532640
|
2.118
|
|
HPN
|
hepsin
|
|
chr19_+_41903723
|
2.118
|
|
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide
|
|
chr12_+_56109851
|
2.116
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr1_+_9599520
|
2.103
|
NM_032315
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr17_-_7197896
|
2.101
|
|
YBX2
|
Y box binding protein 2
|
|
chr12_-_103310896
|
2.083
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr15_-_100882105
|
2.079
|
NM_139057
|
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17
|
|
chr6_-_35656610
|
2.071
|
|
FKBP5
|
FK506 binding protein 5
|
|
chr17_+_46018888
|
2.064
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr19_+_13056671
|
2.059
|
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr12_-_103311053
|
2.054
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr22_-_50699938
|
2.047
|
NM_002969
|
MAPK12
|
mitogen-activated protein kinase 12
|
|
chr16_-_81129882
|
2.040
|
NM_004483
|
GCSH
|
glycine cleavage system protein H (aminomethyl carrier)
|
|
chr8_+_22224786
|
2.037
|
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr17_+_67410825
|
2.022
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr19_+_4791757
|
2.017
|
|
FEM1A
|
fem-1 homolog a (C. elegans)
|
|
chr19_+_13056607
|
2.009
|
NM_005053
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr2_-_200715895
|
1.995
|
|
FONG
|
uncharacterized LOC348751
|
|
chrX_-_114797017
|
1.976
|
|
|
|
|
chr13_+_113951461
|
1.976
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr14_-_24658010
|
1.972
|
NM_024658
|
IPO4
|
importin 4
|
|
chr1_-_167905214
|
1.972
|
|
BRP44
|
brain protein 44
|
|
chrY_+_21729214
|
1.968
|
|
TXLNG2P
|
taxilin gamma 2, pseudogene
|
|
chr11_+_69455837
|
1.968
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr7_-_27205148
|
1.954
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr19_-_12886421
|
1.951
|
NM_001100176
NM_013312
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr10_-_35103894
|
1.951
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr11_+_120081669
|
1.946
|
NM_178507
|
OAF
|
OAF homolog (Drosophila)
|
|
chr8_-_124553253
|
1.941
|
|
FBXO32
|
F-box protein 32
|
|
chr10_-_76995685
|
1.941
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr2_-_101767714
|
1.936
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr1_-_151798552
|
1.936
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr16_-_1876792
|
1.934
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr1_+_32712817
|
1.933
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chr19_+_39971448
|
1.928
|
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae)
|
|
chr6_+_31982538
|
1.928
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr15_+_96875793
|
1.925
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr8_-_9009050
|
1.919
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr16_-_1877140
|
1.903
|
NM_001040427
NM_005326
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr19_-_39466290
|
1.895
|
NM_024907
|
FBXO17
|
F-box protein 17
|
|
chr11_-_57283145
|
1.894
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr12_-_63546589
|
1.885
|
NM_000706
|
AVPR1A
|
arginine vasopressin receptor 1A
|
|
chr19_+_39971496
|
1.884
|
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae)
|
|
chr11_+_7535213
|
1.883
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr11_+_77774902
|
1.880
|
NM_003251
|
THRSP
|
thyroid hormone responsive
|
|
chrX_+_73641084
|
1.879
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr3_-_15900928
|
1.873
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr3_+_183967414
|
1.873
|
NM_014693
NM_032331
|
ECE2
|
endothelin converting enzyme 2
|
|
chr7_+_16793320
|
1.868
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr19_-_48018277
|
1.860
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|