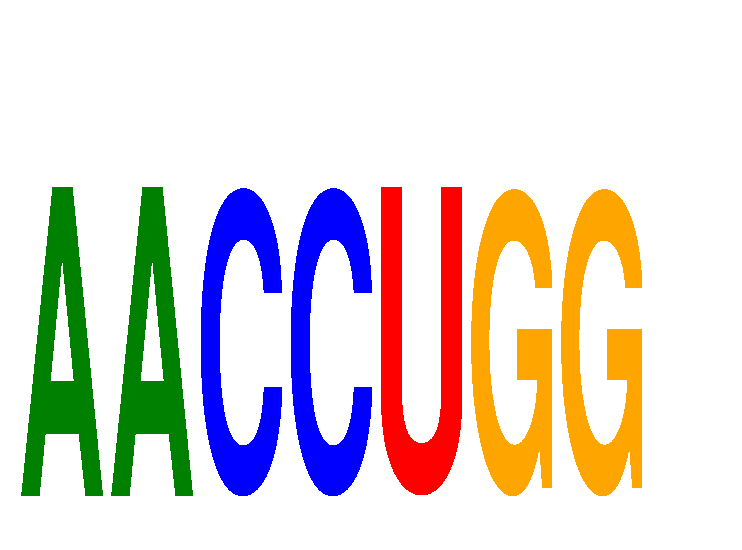Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for AACCUGG
Z-value: 0.11
miRNA associated with seed AACCUGG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-490-3p
|
MIMAT0002806 |
Activity profile of AACCUGG motif
Sorted Z-values of AACCUGG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_55102917 | 0.25 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr2_+_171571827 | 0.18 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr8_-_139509065 | 0.13 |
ENST00000395297.1
|
FAM135B
|
family with sequence similarity 135, member B |
| chr4_+_184020398 | 0.11 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr5_+_109025067 | 0.10 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr16_+_1203194 | 0.10 |
ENST00000348261.5
ENST00000358590.4 |
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr11_-_115375107 | 0.09 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr3_-_189838670 | 0.09 |
ENST00000319332.5
|
LEPREL1
|
leprecan-like 1 |
| chr9_-_38069208 | 0.08 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr1_-_230561475 | 0.08 |
ENST00000391860.1
|
PGBD5
|
piggyBac transposable element derived 5 |
| chr11_+_125462690 | 0.07 |
ENST00000392708.4
ENST00000529196.1 ENST00000531491.1 |
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr7_-_139876812 | 0.06 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr4_-_113437328 | 0.06 |
ENST00000313341.3
|
NEUROG2
|
neurogenin 2 |
| chrX_+_123095155 | 0.05 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chr7_+_100797678 | 0.05 |
ENST00000337619.5
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr8_-_103876965 | 0.05 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr15_-_27018175 | 0.04 |
ENST00000311550.5
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chrX_-_129402857 | 0.04 |
ENST00000447817.1
ENST00000370978.4 |
ZNF280C
|
zinc finger protein 280C |
| chr19_+_4007644 | 0.04 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr8_+_41435696 | 0.03 |
ENST00000396987.3
ENST00000519853.1 |
AGPAT6
|
1-acylglycerol-3-phosphate O-acyltransferase 6 |
| chr8_-_68255912 | 0.03 |
ENST00000262215.3
ENST00000519436.1 |
ARFGEF1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr9_-_123964114 | 0.03 |
ENST00000373840.4
|
RAB14
|
RAB14, member RAS oncogene family |
| chr22_+_25465786 | 0.03 |
ENST00000401395.1
|
KIAA1671
|
KIAA1671 |
| chr18_+_42260861 | 0.03 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr11_+_125439298 | 0.02 |
ENST00000278903.6
ENST00000343678.4 ENST00000524723.1 ENST00000527842.2 |
EI24
|
etoposide induced 2.4 |
| chr4_+_48343339 | 0.02 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr4_+_71768043 | 0.02 |
ENST00000502869.1
ENST00000309395.2 ENST00000396051.2 |
MOB1B
|
MOB kinase activator 1B |
| chr17_-_27621125 | 0.02 |
ENST00000579665.1
ENST00000225388.4 |
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr2_+_178077477 | 0.02 |
ENST00000411529.2
ENST00000435711.1 |
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr7_+_77166592 | 0.02 |
ENST00000248594.6
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr15_+_52121822 | 0.01 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr14_+_57735614 | 0.01 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr11_+_63706444 | 0.01 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr13_-_41240717 | 0.01 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr17_-_48785216 | 0.01 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr11_-_129062093 | 0.01 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_-_91006461 | 0.00 |
ENST00000257749.4
ENST00000343122.3 ENST00000406998.2 ENST00000453877.1 |
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr17_-_1303462 | 0.00 |
ENST00000573026.1
ENST00000575977.1 ENST00000571732.1 ENST00000264335.8 |
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
| chr1_+_93811438 | 0.00 |
ENST00000370272.4
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
| chr17_-_71228357 | 0.00 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of AACCUGG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |