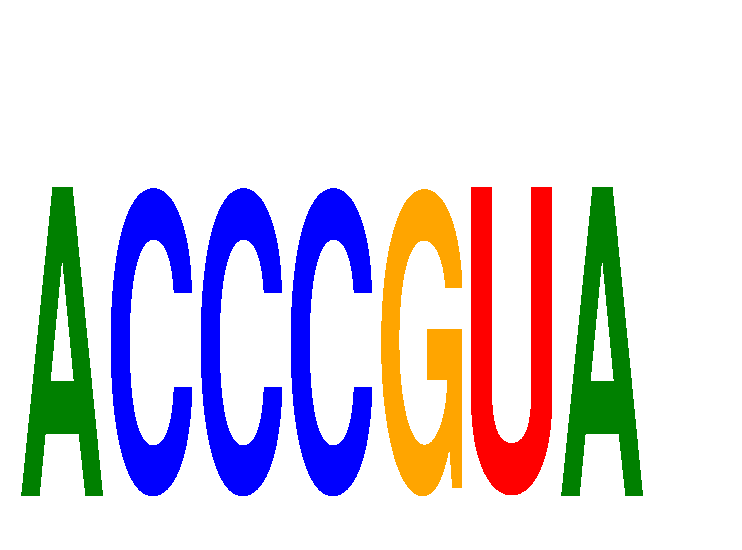Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for ACCCGUA
Z-value: 0.01
miRNA associated with seed ACCCGUA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-99a-5p
|
MIMAT0000097 |
|
hsa-miR-99b-5p
|
MIMAT0000689 |
|
hsa-miR-100-5p
|
MIMAT0000098 |
Activity profile of ACCCGUA motif
Sorted Z-values of ACCCGUA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_141645645 | 0.08 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr3_+_32859510 | 0.08 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr3_+_152017181 | 0.07 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr16_-_80838195 | 0.05 |
ENST00000570137.2
|
CDYL2
|
chromodomain protein, Y-like 2 |
| chr12_-_57030115 | 0.04 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr2_-_72375167 | 0.04 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr8_+_126442563 | 0.03 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr22_+_30279144 | 0.03 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr17_-_4046257 | 0.03 |
ENST00000381638.2
|
ZZEF1
|
zinc finger, ZZ-type with EF-hand domain 1 |
| chr6_-_90121938 | 0.03 |
ENST00000369415.4
|
RRAGD
|
Ras-related GTP binding D |
| chr7_-_27135591 | 0.02 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr2_+_12857015 | 0.02 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr2_+_28974668 | 0.02 |
ENST00000296122.6
ENST00000395366.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_-_157108130 | 0.02 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr1_+_244816237 | 0.02 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr10_-_35930219 | 0.02 |
ENST00000374694.1
|
FZD8
|
frizzled family receptor 8 |
| chr7_+_30323923 | 0.01 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
| chr4_+_144434584 | 0.01 |
ENST00000283131.3
|
SMARCA5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr11_+_86748863 | 0.01 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr2_+_86947296 | 0.00 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr1_+_26560676 | 0.00 |
ENST00000451429.2
ENST00000252992.4 |
CEP85
|
centrosomal protein 85kDa |
| chr4_-_102268628 | 0.00 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_+_65210772 | 0.00 |
ENST00000371072.4
ENST00000294428.3 |
RAVER2
|
ribonucleoprotein, PTB-binding 2 |
| chr19_-_4066890 | 0.00 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
Network of associatons between targets according to the STRING database.
First level regulatory network of ACCCGUA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:0090625 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |