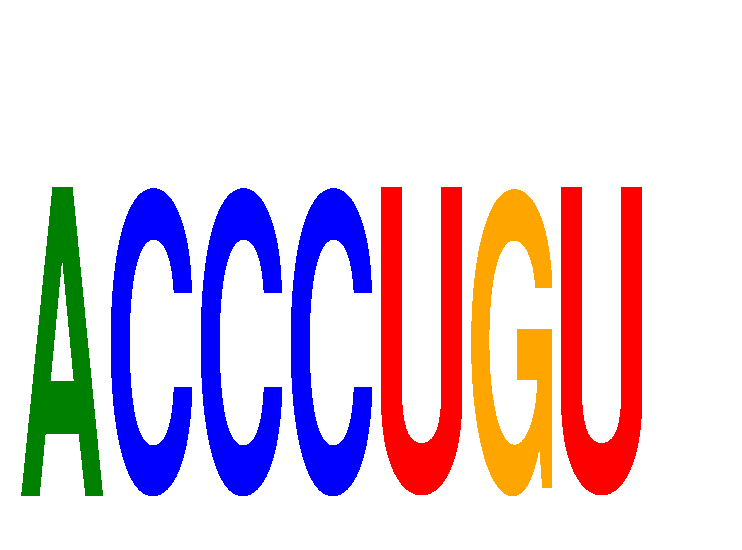Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for ACCCUGU
Z-value: 0.03
miRNA associated with seed ACCCUGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-10a-5p
|
MIMAT0000253 |
|
hsa-miR-10b-5p
|
MIMAT0000254 |
Activity profile of ACCCUGU motif
Sorted Z-values of ACCCUGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_45696253 | 2.04 |
ENST00000303230.4
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr12_+_125811162 | 1.85 |
ENST00000299308.3
|
TMEM132B
|
transmembrane protein 132B |
| chr5_-_160973649 | 1.58 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr1_-_217262969 | 1.58 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr3_-_9595480 | 1.57 |
ENST00000287585.6
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4 |
| chr2_-_2334888 | 1.56 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr8_+_1449532 | 1.49 |
ENST00000421627.2
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2 |
| chr7_-_44365020 | 1.46 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr4_-_100575781 | 1.38 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr1_+_57110972 | 1.38 |
ENST00000371244.4
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr2_+_166326157 | 1.34 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr5_+_167181917 | 1.33 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr6_+_107811162 | 1.29 |
ENST00000317357.5
|
SOBP
|
sine oculis binding protein homolog (Drosophila) |
| chr5_+_143584814 | 1.28 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr4_+_47033345 | 1.25 |
ENST00000295454.3
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr17_+_4487294 | 1.23 |
ENST00000338859.4
|
SMTNL2
|
smoothelin-like 2 |
| chr9_-_23821273 | 1.22 |
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr4_-_53525406 | 1.16 |
ENST00000451218.2
ENST00000441222.3 |
USP46
|
ubiquitin specific peptidase 46 |
| chr1_+_110082487 | 1.14 |
ENST00000527748.1
|
GPR61
|
G protein-coupled receptor 61 |
| chr10_-_62149433 | 1.14 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_-_47812321 | 1.12 |
ENST00000357362.3
ENST00000486952.2 ENST00000426342.1 |
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chrX_+_28605516 | 1.10 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr3_-_179754706 | 1.01 |
ENST00000465751.1
ENST00000467460.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr17_+_34058639 | 0.97 |
ENST00000268864.3
|
RASL10B
|
RAS-like, family 10, member B |
| chr5_-_124080203 | 0.96 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr14_+_33408449 | 0.92 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr7_+_20370746 | 0.86 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr19_-_55954230 | 0.84 |
ENST00000376325.4
|
SHISA7
|
shisa family member 7 |
| chr14_+_85996471 | 0.79 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr15_-_65715401 | 0.77 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr6_-_11044509 | 0.75 |
ENST00000354666.3
|
ELOVL2
|
ELOVL fatty acid elongase 2 |
| chr16_-_70719925 | 0.73 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr9_+_77112244 | 0.70 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr4_-_77135046 | 0.64 |
ENST00000264896.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr19_+_13135386 | 0.62 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr14_-_48143999 | 0.62 |
ENST00000439988.3
|
MDGA2
|
MAM domain-containing glycosylphosphatidylinositol anchor protein 2 |
| chr19_+_10527449 | 0.59 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr4_+_134070439 | 0.59 |
ENST00000264360.5
|
PCDH10
|
protocadherin 10 |
| chr3_+_85008089 | 0.58 |
ENST00000383699.3
|
CADM2
|
cell adhesion molecule 2 |
| chr12_-_14956396 | 0.56 |
ENST00000535328.1
ENST00000261167.2 |
WBP11
|
WW domain binding protein 11 |
| chr4_-_152147579 | 0.56 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr15_-_56209306 | 0.55 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr1_-_9970227 | 0.55 |
ENST00000377263.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr2_-_37193606 | 0.53 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr10_+_103113802 | 0.51 |
ENST00000370187.3
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr9_+_112403088 | 0.49 |
ENST00000448454.2
|
PALM2
|
paralemmin 2 |
| chr11_+_66025167 | 0.48 |
ENST00000394067.2
ENST00000316924.5 ENST00000421552.1 ENST00000394078.1 |
KLC2
|
kinesin light chain 2 |
| chr22_-_17602200 | 0.47 |
ENST00000399875.1
|
CECR6
|
cat eye syndrome chromosome region, candidate 6 |
| chr1_-_85156216 | 0.47 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_+_22889953 | 0.46 |
ENST00000374644.4
ENST00000166244.3 ENST00000538803.1 |
EPHA8
|
EPH receptor A8 |
| chr10_-_128994422 | 0.45 |
ENST00000522781.1
|
FAM196A
|
family with sequence similarity 196, member A |
| chr11_-_115375107 | 0.45 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr4_-_66536057 | 0.44 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr2_-_69870835 | 0.41 |
ENST00000409085.4
ENST00000406297.3 |
AAK1
|
AP2 associated kinase 1 |
| chr11_+_20620946 | 0.41 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr9_-_127533519 | 0.41 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr9_+_129567282 | 0.41 |
ENST00000449886.1
ENST00000373464.4 ENST00000450858.1 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr8_-_41655107 | 0.41 |
ENST00000347528.4
ENST00000289734.7 ENST00000379758.2 ENST00000396945.1 ENST00000396942.1 ENST00000352337.4 |
ANK1
|
ankyrin 1, erythrocytic |
| chr4_+_184020398 | 0.40 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr2_+_97454321 | 0.40 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr6_-_146135880 | 0.39 |
ENST00000237281.4
|
FBXO30
|
F-box protein 30 |
| chr19_+_3880581 | 0.35 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr16_+_69599861 | 0.32 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr6_+_117002339 | 0.31 |
ENST00000413340.1
ENST00000368564.1 ENST00000356348.1 |
KPNA5
|
karyopherin alpha 5 (importin alpha 6) |
| chr1_+_93913713 | 0.31 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr14_+_23775971 | 0.30 |
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2 |
| chr11_+_18344106 | 0.28 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr8_-_4852218 | 0.27 |
ENST00000400186.3
ENST00000602723.1 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr10_+_72972281 | 0.26 |
ENST00000335350.6
|
UNC5B
|
unc-5 homolog B (C. elegans) |
| chr17_+_46125707 | 0.25 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr11_+_113930291 | 0.25 |
ENST00000335953.4
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr4_+_154125565 | 0.23 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr2_-_166060571 | 0.23 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr5_+_63461642 | 0.22 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr22_+_39898325 | 0.22 |
ENST00000325301.2
ENST00000404569.1 |
MIEF1
|
mitochondrial elongation factor 1 |
| chr10_-_79686284 | 0.22 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr6_+_116421976 | 0.21 |
ENST00000319550.4
ENST00000419791.1 |
NT5DC1
|
5'-nucleotidase domain containing 1 |
| chr20_+_55204351 | 0.21 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr8_+_22857048 | 0.19 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chrX_-_54384425 | 0.19 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr8_-_30891078 | 0.18 |
ENST00000339382.2
ENST00000475541.1 |
PURG
|
purine-rich element binding protein G |
| chr5_+_56205081 | 0.17 |
ENST00000285947.2
ENST00000541720.1 |
SETD9
|
SET domain containing 9 |
| chr1_+_224301787 | 0.16 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr6_+_71377473 | 0.16 |
ENST00000370452.3
ENST00000316999.5 ENST00000370455.3 |
SMAP1
|
small ArfGAP 1 |
| chrX_-_46618490 | 0.16 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr3_+_14989076 | 0.15 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr12_+_4918342 | 0.14 |
ENST00000280684.3
ENST00000433855.1 |
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr17_-_48207157 | 0.13 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr15_-_61521495 | 0.12 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr10_+_180987 | 0.12 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr17_-_49198216 | 0.12 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr18_-_33647487 | 0.11 |
ENST00000590898.1
ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr9_+_100263912 | 0.11 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr13_-_29069232 | 0.11 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr21_+_45285050 | 0.10 |
ENST00000291572.8
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr12_+_69864129 | 0.10 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr17_-_73775839 | 0.09 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr16_-_85722530 | 0.09 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr2_+_64681219 | 0.09 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr2_-_206950781 | 0.08 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr10_-_30348439 | 0.08 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462 |
| chr17_-_4167142 | 0.07 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr4_-_146859623 | 0.06 |
ENST00000379448.4
ENST00000513320.1 |
ZNF827
|
zinc finger protein 827 |
| chr17_+_44928946 | 0.06 |
ENST00000290015.2
ENST00000393461.2 |
WNT9B
|
wingless-type MMTV integration site family, member 9B |
| chr19_-_14316980 | 0.05 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr6_+_15246501 | 0.05 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr7_-_138666053 | 0.05 |
ENST00000440172.1
ENST00000422774.1 |
KIAA1549
|
KIAA1549 |
| chr2_+_215275771 | 0.05 |
ENST00000312504.5
ENST00000427124.1 |
VWC2L
|
von Willebrand factor C domain containing protein 2-like |
| chr18_+_43914159 | 0.04 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr19_+_13229126 | 0.04 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr3_-_9994021 | 0.04 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr1_+_89990431 | 0.04 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr17_+_45727204 | 0.03 |
ENST00000290158.4
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr7_+_94139105 | 0.03 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr1_+_169337172 | 0.03 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr2_-_164592497 | 0.03 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr3_+_183873098 | 0.02 |
ENST00000313143.3
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr9_-_115983641 | 0.02 |
ENST00000238256.3
|
FKBP15
|
FK506 binding protein 15, 133kDa |
| chr4_-_111119804 | 0.02 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_-_75173785 | 0.01 |
ENST00000535178.1
ENST00000372921.5 ENST00000372919.4 |
ANXA7
|
annexin A7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ACCCUGU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.2 | 2.7 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 1.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.6 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 0.8 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 2.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.5 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 1.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.6 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.1 | 0.3 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 1.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 1.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.4 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.6 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.6 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 1.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.7 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.4 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.6 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.9 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.7 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.3 | 0.9 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.5 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 1.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.4 | 2.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.3 | 1.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 1.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 1.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.4 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 1.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.1 | 0.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.2 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |