Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for ALX1_ARX
Z-value: 0.26
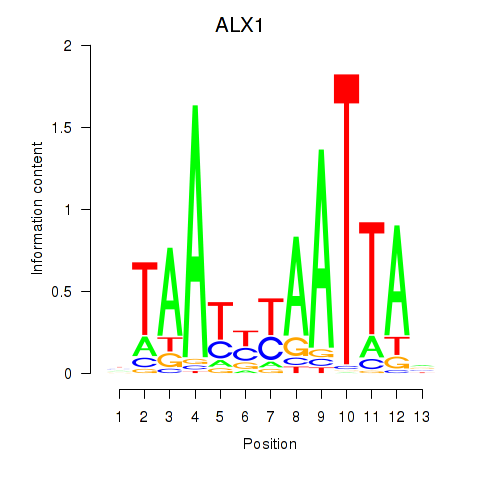
Transcription factors associated with ALX1_ARX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX1
|
ENSG00000180318.3 | ALX homeobox 1 |
|
ARX
|
ENSG00000004848.6 | aristaless related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARX | hg19_v2_chrX_-_25034065_25034088 | 0.09 | 6.3e-01 | Click! |
| ALX1 | hg19_v2_chr12_+_85673868_85673885 | -0.07 | 7.1e-01 | Click! |
Activity profile of ALX1_ARX motif
Sorted Z-values of ALX1_ARX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_95236551 | 4.62 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr14_+_22631122 | 4.61 |
ENST00000390458.3
|
TRAV29DV5
|
T cell receptor alpha variable 29/delta variable 5 (gene/pseudogene) |
| chr3_-_27763803 | 2.76 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr3_-_27764190 | 2.47 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr5_+_96840389 | 2.39 |
ENST00000504012.1
|
RP11-1E3.1
|
RP11-1E3.1 |
| chr1_-_151148492 | 2.25 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr1_-_151148442 | 2.23 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr6_-_66417107 | 2.01 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr3_-_3151664 | 1.96 |
ENST00000256452.3
ENST00000311981.8 ENST00000430514.2 ENST00000456302.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr12_+_54410664 | 1.91 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr3_-_157824292 | 1.75 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr1_-_92371839 | 1.30 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr8_-_57472154 | 1.25 |
ENST00000499425.1
ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968
|
long intergenic non-protein coding RNA 968 |
| chr17_+_61151306 | 1.19 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chrX_-_19817869 | 1.18 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr3_+_138340049 | 1.13 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr8_-_57472137 | 1.11 |
ENST00000519144.1
|
LINC00968
|
long intergenic non-protein coding RNA 968 |
| chr2_-_89160770 | 1.10 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr1_+_81106951 | 1.10 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chrX_+_107288197 | 1.09 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_107288280 | 1.08 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr8_+_86999516 | 1.07 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr7_-_115799942 | 1.06 |
ENST00000484212.1
|
TFEC
|
transcription factor EC |
| chrX_+_107288239 | 1.05 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_+_176994408 | 0.94 |
ENST00000429017.1
ENST00000313173.4 ENST00000544999.1 |
HOXD8
|
homeobox D8 |
| chr10_-_29923893 | 0.93 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr2_-_89161432 | 0.93 |
ENST00000390242.2
|
IGKJ1
|
immunoglobulin kappa joining 1 |
| chr10_-_75226166 | 0.91 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr2_-_145188137 | 0.87 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr13_+_74805561 | 0.83 |
ENST00000419499.1
|
LINC00402
|
long intergenic non-protein coding RNA 402 |
| chr7_+_30589829 | 0.83 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr12_-_57873329 | 0.80 |
ENST00000424809.2
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr6_-_32908792 | 0.79 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_-_8964580 | 0.79 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr6_-_32908765 | 0.79 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr12_-_53601055 | 0.77 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr4_-_36245561 | 0.76 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_121659052 | 0.75 |
ENST00000512105.1
|
CTD-2544H17.1
|
CTD-2544H17.1 |
| chr1_+_170632250 | 0.75 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr7_+_93535817 | 0.74 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr8_-_57472049 | 0.73 |
ENST00000523786.1
ENST00000521483.1 |
LINC00968
|
long intergenic non-protein coding RNA 968 |
| chr12_-_53601000 | 0.73 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr14_-_83262540 | 0.73 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr11_+_128563652 | 0.73 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_-_15332665 | 0.72 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr12_-_91546926 | 0.72 |
ENST00000550758.1
|
DCN
|
decorin |
| chr18_-_44181442 | 0.71 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr1_-_178838404 | 0.71 |
ENST00000444255.1
|
ANGPTL1
|
angiopoietin-like 1 |
| chr2_-_145278475 | 0.68 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_206557366 | 0.67 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr12_-_57873631 | 0.65 |
ENST00000393791.3
ENST00000356411.2 ENST00000552249.1 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr2_+_36923830 | 0.64 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr1_+_192127578 | 0.64 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr14_-_52436247 | 0.62 |
ENST00000597846.1
|
AL358333.1
|
HCG2013195; Uncharacterized protein |
| chr12_+_14572070 | 0.61 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_36923933 | 0.60 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chr12_-_89746264 | 0.57 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr6_-_133055896 | 0.57 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr3_+_151591422 | 0.57 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr10_-_50396425 | 0.54 |
ENST00000374148.1
|
C10orf128
|
chromosome 10 open reading frame 128 |
| chr17_+_44588877 | 0.54 |
ENST00000576629.1
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr12_-_92539614 | 0.54 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr16_+_68119247 | 0.54 |
ENST00000575270.1
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr12_-_89746173 | 0.53 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr8_+_77318769 | 0.53 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr19_+_50016610 | 0.52 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr2_+_119699864 | 0.52 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr16_+_68119324 | 0.52 |
ENST00000349223.5
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr16_+_68119440 | 0.52 |
ENST00000346183.3
ENST00000329524.4 |
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr10_-_50396407 | 0.51 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr2_+_119699742 | 0.51 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr8_-_122653630 | 0.50 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr12_+_122241928 | 0.50 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chr4_-_68829226 | 0.49 |
ENST00000396188.2
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr1_+_186265399 | 0.49 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr14_-_75083313 | 0.49 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr16_+_53469525 | 0.48 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr4_+_69313145 | 0.48 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr16_+_22518495 | 0.48 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chrX_-_11284095 | 0.47 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr1_+_186798073 | 0.47 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr6_-_133055815 | 0.47 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr9_+_44867571 | 0.46 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr2_-_89161064 | 0.46 |
ENST00000390241.2
|
IGKJ2
|
immunoglobulin kappa joining 2 |
| chr8_+_19759228 | 0.46 |
ENST00000520959.1
|
LPL
|
lipoprotein lipase |
| chr11_+_108094786 | 0.45 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chr3_-_191000172 | 0.45 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr2_-_145277882 | 0.45 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr10_+_69865866 | 0.45 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr2_+_36923901 | 0.45 |
ENST00000457137.2
|
VIT
|
vitrin |
| chr15_+_42066632 | 0.45 |
ENST00000457542.2
ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr10_-_31146615 | 0.44 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr12_-_52946923 | 0.43 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr3_+_121774202 | 0.42 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr1_+_158978768 | 0.42 |
ENST00000447473.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr8_-_6420930 | 0.40 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr4_-_143227088 | 0.40 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr12_-_118796910 | 0.40 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr17_-_60885645 | 0.40 |
ENST00000544856.2
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr11_-_111649015 | 0.40 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr5_+_137722255 | 0.39 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr4_-_143226979 | 0.37 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr6_+_34204642 | 0.36 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chrX_+_77166172 | 0.36 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr6_+_15401075 | 0.35 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr17_+_39261584 | 0.35 |
ENST00000391415.1
|
KRTAP4-9
|
keratin associated protein 4-9 |
| chr1_+_154401791 | 0.35 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr11_-_96076334 | 0.35 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr10_-_50396357 | 0.35 |
ENST00000453436.1
ENST00000474718.1 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr4_-_68829144 | 0.35 |
ENST00000508048.1
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr22_-_30642728 | 0.35 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr7_-_55620433 | 0.35 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr9_-_95298254 | 0.35 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr17_-_60885659 | 0.33 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr10_+_97803151 | 0.32 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr17_+_74463650 | 0.32 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr11_-_16419067 | 0.32 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr3_-_186262166 | 0.31 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr17_-_60885700 | 0.31 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr10_-_102989551 | 0.31 |
ENST00000370193.2
|
LBX1
|
ladybird homeobox 1 |
| chr17_-_38956205 | 0.31 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr6_-_26199499 | 0.30 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr12_+_1099675 | 0.30 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr6_-_150067696 | 0.30 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr1_-_178840157 | 0.29 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr2_+_58655461 | 0.29 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr5_-_39270725 | 0.29 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr16_-_21875424 | 0.29 |
ENST00000541674.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr12_-_125052010 | 0.28 |
ENST00000458234.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr1_+_62439037 | 0.28 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr3_+_138340067 | 0.27 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_-_119759795 | 0.27 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chrX_+_11311533 | 0.27 |
ENST00000380714.3
ENST00000380712.3 ENST00000348912.4 |
AMELX
|
amelogenin, X-linked |
| chr7_-_33842742 | 0.25 |
ENST00000420185.1
ENST00000440034.1 |
RP11-89N17.4
|
RP11-89N17.4 |
| chr7_+_135710494 | 0.25 |
ENST00000440744.2
|
AC024084.1
|
AC024084.1 |
| chr2_+_54683419 | 0.25 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr18_-_52989217 | 0.25 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr9_+_135854091 | 0.24 |
ENST00000450530.1
ENST00000534944.1 |
GFI1B
|
growth factor independent 1B transcription repressor |
| chrX_+_39868501 | 0.24 |
ENST00000447651.1
|
AC092198.1
|
AC092198.1 |
| chr2_+_29001711 | 0.24 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr6_-_137314371 | 0.24 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr14_+_57671888 | 0.24 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr12_-_15815626 | 0.24 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr8_-_62602327 | 0.24 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr18_-_52989525 | 0.23 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr3_+_12329358 | 0.23 |
ENST00000309576.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr3_+_12329397 | 0.23 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr3_-_101039402 | 0.22 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr10_-_99030395 | 0.22 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr6_-_26199471 | 0.22 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr4_+_76649753 | 0.22 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr11_-_58035732 | 0.22 |
ENST00000395079.2
|
OR10W1
|
olfactory receptor, family 10, subfamily W, member 1 |
| chrX_+_16668278 | 0.22 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr3_-_124774802 | 0.22 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr5_-_59783882 | 0.22 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_118628350 | 0.22 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr14_-_81425828 | 0.22 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr4_-_103940791 | 0.22 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr15_-_98836406 | 0.22 |
ENST00000560360.1
|
CTD-2544M6.1
|
CTD-2544M6.1 |
| chr5_+_68860949 | 0.21 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr5_-_60411762 | 0.21 |
ENST00000594278.1
|
AC008498.1
|
AC008498.1 |
| chr17_-_71228357 | 0.21 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr14_-_57197224 | 0.21 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr5_+_140227357 | 0.21 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr3_-_195310802 | 0.21 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr14_+_22951993 | 0.20 |
ENST00000390485.1
|
TRAJ53
|
T cell receptor alpha joining 53 |
| chr14_-_78083112 | 0.20 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr2_+_161993465 | 0.20 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr5_+_91378410 | 0.19 |
ENST00000507217.1
|
RP11-348J24.2
|
RP11-348J24.2 |
| chr3_-_32544900 | 0.19 |
ENST00000205636.3
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chrX_+_107037451 | 0.19 |
ENST00000372379.2
|
NCBP2L
|
nuclear cap binding protein subunit 2-like |
| chr6_-_170893268 | 0.19 |
ENST00000538195.1
|
PDCD2
|
programmed cell death 2 |
| chr6_-_111804905 | 0.19 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr1_+_244214577 | 0.18 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr9_+_123884038 | 0.18 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr16_+_24549014 | 0.18 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr2_-_145275228 | 0.18 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr9_+_88556444 | 0.18 |
ENST00000376040.1
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr5_-_134735568 | 0.18 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr2_+_161993412 | 0.18 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr21_-_43346790 | 0.17 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr1_+_158323755 | 0.17 |
ENST00000368157.1
ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E
|
CD1e molecule |
| chr2_-_145275828 | 0.17 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_118955500 | 0.17 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr4_-_83769996 | 0.17 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr12_+_59989918 | 0.17 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr7_-_93520259 | 0.17 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr15_+_43477580 | 0.16 |
ENST00000356633.5
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chrX_+_36254051 | 0.16 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr2_+_102413726 | 0.16 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_-_118628315 | 0.16 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr5_+_145826867 | 0.16 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr12_+_10163231 | 0.16 |
ENST00000396502.1
ENST00000338896.5 |
CLEC12B
|
C-type lectin domain family 12, member B |
| chr1_-_154600421 | 0.16 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr4_+_95128996 | 0.16 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr10_+_97709725 | 0.16 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr6_+_3259122 | 0.15 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr2_-_31440377 | 0.15 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr8_-_61880248 | 0.15 |
ENST00000525556.1
|
AC022182.3
|
AC022182.3 |
| chr3_+_130569429 | 0.15 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX1_ARX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.5 | 4.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.5 | 1.6 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.4 | 4.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 2.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.3 | 1.5 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.3 | 2.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 1.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 1.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.5 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.2 | 0.7 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 1.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.4 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.5 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.5 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.3 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.4 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.5 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.4 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 1.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.6 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.4 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 2.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 2.0 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.3 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.9 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 1.6 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 1.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.4 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.9 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.7 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 1.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.3 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 1.0 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.3 | 1.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 4.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.4 | 2.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 1.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.8 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 3.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 5.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 5.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 2.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 2.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.0 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0005262 | calcium channel activity(GO:0005262) calcium ion transmembrane transporter activity(GO:0015085) |
| 0.0 | 0.2 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 5.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 2.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |