Project
Illumina Body Map 2, young vs old
Navigation
Downloads
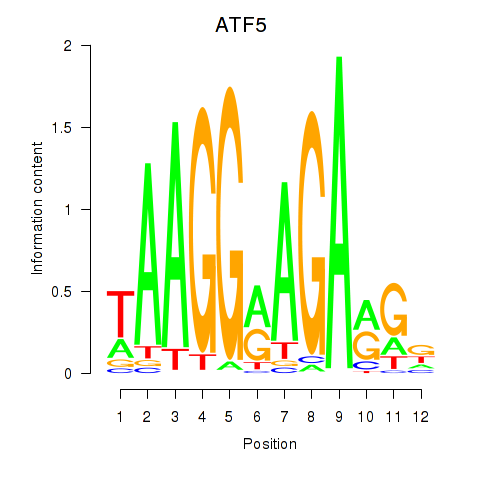
Results for ATF5
Z-value: 2.00
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF5 | hg19_v2_chr19_+_50433476_50433510 | -0.33 | 6.8e-02 | Click! |
Activity profile of ATF5 motif
Sorted Z-values of ATF5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_52612705 | 6.28 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr20_-_52612468 | 4.98 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr14_+_23846210 | 4.00 |
ENST00000339180.4
ENST00000342473.4 ENST00000397227.3 ENST00000555731.1 |
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr14_+_23846328 | 3.52 |
ENST00000382809.2
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr14_+_23845995 | 3.44 |
ENST00000359320.3
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr5_+_175223313 | 2.86 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr5_+_156887027 | 2.79 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr2_-_87017985 | 2.77 |
ENST00000352580.3
|
CD8A
|
CD8a molecule |
| chr2_-_87018784 | 2.69 |
ENST00000283635.3
ENST00000538832.1 |
CD8A
|
CD8a molecule |
| chr14_+_92789498 | 2.42 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr7_-_3083472 | 2.31 |
ENST00000356408.3
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr4_-_176733377 | 2.25 |
ENST00000505375.1
|
GPM6A
|
glycoprotein M6A |
| chr5_-_39219705 | 2.24 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr5_-_148442584 | 2.11 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr5_-_39219641 | 2.07 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr5_-_148442687 | 2.04 |
ENST00000515425.1
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr5_+_110559603 | 2.00 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr22_-_37215523 | 1.98 |
ENST00000216200.5
|
PVALB
|
parvalbumin |
| chr18_-_3845321 | 1.92 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_+_159442878 | 1.92 |
ENST00000307765.5
ENST00000423548.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr8_-_131028782 | 1.87 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr3_-_62861012 | 1.81 |
ENST00000357948.3
ENST00000383710.4 |
CADPS
|
Ca++-dependent secretion activator |
| chr7_-_3083573 | 1.73 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr2_+_191334212 | 1.61 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr6_-_62996066 | 1.60 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr14_-_36988882 | 1.60 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chrX_-_13835147 | 1.58 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr4_+_159443024 | 1.54 |
ENST00000448688.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr5_+_110559312 | 1.53 |
ENST00000508074.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chrX_-_13835461 | 1.52 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr18_-_3845293 | 1.51 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr6_-_31514333 | 1.49 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr3_-_62860878 | 1.47 |
ENST00000283269.9
|
CADPS
|
Ca++-dependent secretion activator |
| chr4_+_159443090 | 1.46 |
ENST00000343542.5
ENST00000470033.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr10_-_104179682 | 1.36 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chrX_-_13835398 | 1.35 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr8_-_131028869 | 1.34 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr10_-_104178857 | 1.34 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr5_-_20575959 | 1.34 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr20_+_58179582 | 1.31 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr16_+_28943260 | 1.31 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr17_-_7108436 | 1.30 |
ENST00000493294.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr2_+_173724771 | 1.26 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr19_-_8642289 | 1.25 |
ENST00000596675.1
ENST00000338257.8 |
MYO1F
|
myosin IF |
| chr16_-_30023615 | 1.25 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chr1_+_202091980 | 1.24 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr22_-_30662828 | 1.20 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr3_+_101504200 | 1.19 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr10_+_111765562 | 1.19 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr2_-_26205550 | 1.18 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
| chr7_-_127672146 | 1.17 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr6_+_72596604 | 1.14 |
ENST00000348717.5
ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr12_+_10124110 | 1.14 |
ENST00000350667.4
|
CLEC12A
|
C-type lectin domain family 12, member A |
| chr19_+_55795493 | 1.10 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr2_-_26205340 | 1.09 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr1_+_36023035 | 1.07 |
ENST00000373253.3
|
NCDN
|
neurochondrin |
| chr5_+_173930676 | 1.07 |
ENST00000504512.1
|
RP11-267A15.1
|
RP11-267A15.1 |
| chr11_-_132813566 | 1.04 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr1_+_36023370 | 1.03 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr19_-_38916839 | 1.03 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr14_+_79746249 | 1.02 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chrX_+_122318318 | 1.01 |
ENST00000371251.1
ENST00000371256.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr3_-_183145765 | 1.00 |
ENST00000473233.1
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr7_+_136553824 | 0.98 |
ENST00000320658.5
ENST00000453373.1 ENST00000397608.3 ENST00000402486.3 ENST00000401861.1 |
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr5_+_82767583 | 0.97 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr1_+_66999799 | 0.97 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr17_+_44790515 | 0.97 |
ENST00000576346.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr19_-_7936344 | 0.96 |
ENST00000599142.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr8_+_79428539 | 0.95 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chrX_-_19817869 | 0.94 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr11_-_132812987 | 0.94 |
ENST00000541867.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr5_+_175223715 | 0.94 |
ENST00000515502.1
|
CPLX2
|
complexin 2 |
| chr3_-_183145873 | 0.93 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr12_+_125811162 | 0.92 |
ENST00000299308.3
|
TMEM132B
|
transmembrane protein 132B |
| chr16_+_50313426 | 0.92 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr11_-_132813627 | 0.92 |
ENST00000374778.4
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chrX_+_122318224 | 0.91 |
ENST00000542149.1
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr19_-_38916802 | 0.90 |
ENST00000587738.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr1_-_183387723 | 0.90 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr5_+_82767487 | 0.89 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr3_-_62860704 | 0.89 |
ENST00000490353.2
|
CADPS
|
Ca++-dependent secretion activator |
| chr15_+_75118888 | 0.88 |
ENST00000395018.4
|
CPLX3
|
complexin 3 |
| chr10_-_7453445 | 0.87 |
ENST00000379713.3
ENST00000397167.1 ENST00000397160.3 |
SFMBT2
|
Scm-like with four mbt domains 2 |
| chr12_-_54778444 | 0.85 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr2_-_26700900 | 0.85 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr14_-_39901618 | 0.85 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chr7_-_37488547 | 0.83 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr10_+_106918686 | 0.83 |
ENST00000393176.2
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr2_-_145278475 | 0.83 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr22_+_37678505 | 0.83 |
ENST00000402997.1
ENST00000405206.3 |
CYTH4
|
cytohesin 4 |
| chrX_+_122318006 | 0.82 |
ENST00000371266.1
ENST00000264357.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr12_-_16761117 | 0.82 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr5_+_82767284 | 0.81 |
ENST00000265077.3
|
VCAN
|
versican |
| chr12_+_10124001 | 0.81 |
ENST00000396507.3
ENST00000304361.4 ENST00000434319.2 |
CLEC12A
|
C-type lectin domain family 12, member A |
| chr9_-_23825956 | 0.80 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr11_-_104480019 | 0.79 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr11_-_126870655 | 0.78 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr7_-_127671674 | 0.77 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr6_+_72596406 | 0.77 |
ENST00000491071.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr19_-_38916822 | 0.76 |
ENST00000586305.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr19_-_44285401 | 0.75 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr12_-_49351303 | 0.74 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr12_-_54778471 | 0.74 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr7_-_115670792 | 0.73 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr16_+_31271274 | 0.73 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr1_+_223354486 | 0.71 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr8_-_21645804 | 0.71 |
ENST00000518077.1
ENST00000517892.1 |
GFRA2
|
GDNF family receptor alpha 2 |
| chr17_+_38337491 | 0.69 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr12_-_16761007 | 0.69 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr2_-_214016314 | 0.67 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr15_+_77287715 | 0.67 |
ENST00000559161.1
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr17_+_33914424 | 0.67 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr15_+_38296918 | 0.67 |
ENST00000557883.2
|
RP11-1008C21.1
|
RP11-1008C21.1 |
| chr9_-_23826298 | 0.66 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr19_+_10531150 | 0.66 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr15_+_77287426 | 0.66 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr7_+_73498118 | 0.65 |
ENST00000336180.2
|
LIMK1
|
LIM domain kinase 1 |
| chr7_-_100061869 | 0.64 |
ENST00000332375.3
|
C7orf61
|
chromosome 7 open reading frame 61 |
| chr17_+_38334501 | 0.63 |
ENST00000541245.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr1_+_206579736 | 0.63 |
ENST00000439126.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr19_-_15090488 | 0.63 |
ENST00000594383.1
ENST00000598504.1 ENST00000597262.1 |
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr2_+_149447783 | 0.62 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr1_-_182641037 | 0.60 |
ENST00000483095.2
|
RGS8
|
regulator of G-protein signaling 8 |
| chr7_-_78400364 | 0.59 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_-_197036289 | 0.58 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr7_-_115670804 | 0.57 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr14_+_79745746 | 0.57 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr1_+_48688357 | 0.57 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr1_+_215179188 | 0.56 |
ENST00000391895.2
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr11_-_126870683 | 0.55 |
ENST00000525704.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chrX_+_131157609 | 0.54 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr5_+_173930710 | 0.54 |
ENST00000511707.1
|
RP11-267A15.1
|
RP11-267A15.1 |
| chr1_+_101361782 | 0.53 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr20_+_54987305 | 0.53 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr11_-_129062093 | 0.52 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr19_+_42773371 | 0.52 |
ENST00000571942.2
|
CIC
|
capicua transcriptional repressor |
| chr12_-_1920886 | 0.52 |
ENST00000536846.2
ENST00000538027.2 ENST00000538450.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr3_-_69129501 | 0.51 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr2_-_100759037 | 0.51 |
ENST00000317233.4
ENST00000423966.1 ENST00000416492.1 |
AFF3
|
AF4/FMR2 family, member 3 |
| chr1_-_182640988 | 0.51 |
ENST00000367556.1
|
RGS8
|
regulator of G-protein signaling 8 |
| chr7_-_102232891 | 0.50 |
ENST00000514917.2
|
RP11-514P8.7
|
RP11-514P8.7 |
| chr9_+_127023704 | 0.46 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr8_-_61193947 | 0.46 |
ENST00000317995.4
|
CA8
|
carbonic anhydrase VIII |
| chr17_-_47287928 | 0.46 |
ENST00000507680.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr20_+_54987168 | 0.46 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr12_+_32654965 | 0.45 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_132009675 | 0.44 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr15_-_75871589 | 0.43 |
ENST00000306726.2
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr17_+_38334242 | 0.42 |
ENST00000436615.3
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr17_+_47287749 | 0.42 |
ENST00000419580.2
|
ABI3
|
ABI family, member 3 |
| chr12_-_25101920 | 0.42 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr12_-_54778244 | 0.42 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr14_+_32547434 | 0.41 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_-_47697387 | 0.41 |
ENST00000371884.2
|
TAL1
|
T-cell acute lymphocytic leukemia 1 |
| chr5_+_138629628 | 0.40 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr8_+_38677850 | 0.39 |
ENST00000518809.1
ENST00000520611.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr14_+_100111447 | 0.38 |
ENST00000330710.5
ENST00000357223.2 |
HHIPL1
|
HHIP-like 1 |
| chr5_-_134788086 | 0.38 |
ENST00000537858.1
|
TIFAB
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chrX_-_19983260 | 0.37 |
ENST00000340625.3
|
CXorf23
|
chromosome X open reading frame 23 |
| chr2_-_113542063 | 0.37 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr8_-_21646290 | 0.37 |
ENST00000519195.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr15_+_78556428 | 0.36 |
ENST00000394855.3
ENST00000489435.2 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr17_+_18761417 | 0.36 |
ENST00000419284.2
ENST00000268835.2 ENST00000412418.1 ENST00000575228.1 ENST00000575102.1 ENST00000536323.1 |
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr1_-_54872059 | 0.36 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr1_+_206137237 | 0.35 |
ENST00000468509.1
ENST00000367129.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr8_-_623547 | 0.35 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr6_+_87865262 | 0.33 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chrX_+_122318113 | 0.33 |
ENST00000371264.3
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr3_+_19988885 | 0.33 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr14_+_32546274 | 0.32 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr5_+_138629417 | 0.32 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr12_-_48597170 | 0.32 |
ENST00000310248.2
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1 |
| chr9_+_103947311 | 0.31 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr17_+_33914276 | 0.31 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr12_-_54652060 | 0.31 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr5_+_138629337 | 0.31 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr1_-_151319318 | 0.31 |
ENST00000436271.1
ENST00000450506.1 ENST00000422595.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr15_+_63481668 | 0.31 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr11_-_33891362 | 0.31 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr8_+_95731904 | 0.30 |
ENST00000522422.1
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chrX_-_44402231 | 0.29 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr19_+_4229495 | 0.28 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr5_+_138629389 | 0.28 |
ENST00000504045.1
ENST00000504311.1 ENST00000502499.1 |
MATR3
|
matrin 3 |
| chr8_+_107738343 | 0.28 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr8_+_107738240 | 0.28 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr1_-_156460391 | 0.28 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr1_-_151319283 | 0.28 |
ENST00000392746.3
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr14_+_32546145 | 0.28 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr17_-_5522731 | 0.27 |
ENST00000576905.1
|
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr18_-_53089723 | 0.27 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr6_-_88411911 | 0.26 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chrX_+_123095155 | 0.26 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chr3_+_23958470 | 0.26 |
ENST00000434031.2
ENST00000413699.1 ENST00000456530.2 |
RPL15
|
ribosomal protein L15 |
| chr4_-_175750364 | 0.25 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
| chr7_-_78400598 | 0.25 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_-_6007787 | 0.24 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr1_-_36022979 | 0.24 |
ENST00000469892.1
ENST00000325722.3 |
KIAA0319L
|
KIAA0319-like |
| chr11_+_58938903 | 0.24 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr17_+_33914460 | 0.24 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr3_+_19988566 | 0.24 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 1.1 | 5.5 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.6 | 4.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.5 | 3.2 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.4 | 1.3 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.4 | 4.5 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 2.0 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.3 | 4.0 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.2 | 4.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 0.4 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.2 | 2.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 0.5 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 6.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 1.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 0.6 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.2 | 1.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.9 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 3.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 1.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 1.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.4 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 3.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.4 | GO:0071288 | cellular response to mercury ion(GO:0071288) positive regulation of mononuclear cell migration(GO:0071677) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 1.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 1.1 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 1.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.8 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 1.9 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 2.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.9 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.3 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.1 | 1.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.8 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 4.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.6 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.1 | 0.4 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.8 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 2.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 2.4 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.3 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 0.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 1.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.4 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.1 | 0.8 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 2.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 3.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.4 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.0 | 3.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.6 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 2.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 1.9 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 2.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 1.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 5.0 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 1.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 3.0 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.6 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.9 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.2 | 9.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 1.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.7 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.3 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 2.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.8 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 3.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 1.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.9 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 2.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 2.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 7.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 12.0 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 5.1 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 2.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.3 | 1.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 2.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.5 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 4.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 3.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.9 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 3.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 5.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.8 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 2.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 4.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.2 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 1.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 1.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 7.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 3.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 2.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 2.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 9.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 3.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.5 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 6.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 4.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 4.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |