Project
Illumina Body Map 2, young vs old
Navigation
Downloads
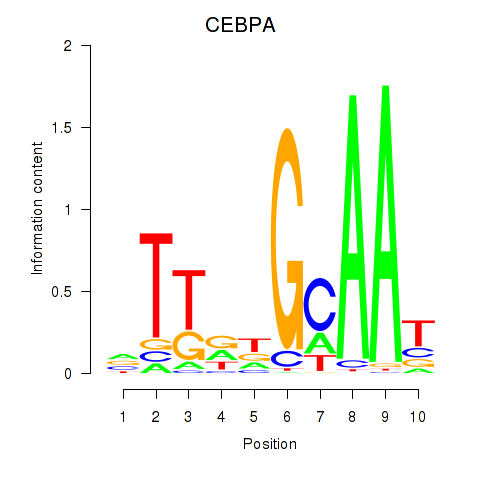
Results for CEBPA
Z-value: 0.07
Transcription factors associated with CEBPA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPA
|
ENSG00000245848.2 | CCAAT enhancer binding protein alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPA | hg19_v2_chr19_-_33793430_33793470 | 0.49 | 4.5e-03 | Click! |
Activity profile of CEBPA motif
Sorted Z-values of CEBPA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_155533787 | 16.77 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr1_-_159684371 | 13.76 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr9_+_130911723 | 13.11 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr9_+_130911770 | 12.01 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr11_-_18258342 | 11.20 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr19_-_4540486 | 10.04 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr16_+_72088376 | 10.01 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr8_-_6837602 | 9.83 |
ENST00000382692.2
|
DEFA1
|
defensin, alpha 1 |
| chr6_-_133055815 | 9.08 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr6_-_133055896 | 8.35 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr8_-_6875778 | 8.29 |
ENST00000535841.1
ENST00000327857.2 |
DEFA1B
DEFA3
|
defensin, alpha 1B defensin, alpha 3, neutrophil-specific |
| chr11_-_102595681 | 8.25 |
ENST00000236826.3
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr12_-_96390108 | 8.19 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr20_-_43883197 | 8.11 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr17_-_7018128 | 7.88 |
ENST00000380952.2
ENST00000254850.7 |
ASGR2
|
asialoglycoprotein receptor 2 |
| chr17_-_7017968 | 7.76 |
ENST00000355035.5
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chr11_+_18287801 | 7.73 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_-_18270182 | 7.67 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr11_+_18287721 | 7.61 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr3_-_120400960 | 7.51 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr11_+_59824060 | 7.43 |
ENST00000395032.2
ENST00000358152.2 |
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr8_-_6795823 | 7.42 |
ENST00000297435.2
|
DEFA4
|
defensin, alpha 4, corticostatin |
| chr12_+_69742121 | 7.28 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr11_+_59824127 | 7.04 |
ENST00000278865.3
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr19_-_48389651 | 6.79 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr11_+_116700600 | 5.91 |
ENST00000227667.3
|
APOC3
|
apolipoprotein C-III |
| chr11_+_116700614 | 5.84 |
ENST00000375345.1
|
APOC3
|
apolipoprotein C-III |
| chr11_-_102595512 | 5.83 |
ENST00000438475.2
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr12_-_10007448 | 5.83 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr1_+_207277590 | 5.76 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr16_+_82090028 | 5.68 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr18_-_61329118 | 5.65 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr11_-_59633951 | 5.61 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr1_+_196743912 | 5.44 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr6_+_31895287 | 5.35 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr1_+_57320437 | 5.34 |
ENST00000361249.3
|
C8A
|
complement component 8, alpha polypeptide |
| chr13_-_46679144 | 5.24 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chrX_-_47489244 | 5.23 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr1_+_207277632 | 5.21 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr13_-_46679185 | 5.08 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr11_-_10590118 | 5.07 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr2_+_218994002 | 4.98 |
ENST00000428565.1
|
CXCR2
|
chemokine (C-X-C motif) receptor 2 |
| chr5_+_147258266 | 4.97 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chr12_-_96390063 | 4.93 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr1_+_196743943 | 4.84 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr1_-_153348067 | 4.80 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr6_+_31895254 | 4.77 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr2_+_234590556 | 4.69 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr15_+_58430368 | 4.68 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr4_-_100242549 | 4.68 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr6_+_31895480 | 4.45 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr19_+_41497178 | 4.32 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr2_+_128177253 | 4.31 |
ENST00000427769.1
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr3_+_186435137 | 4.27 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr21_-_37852359 | 4.24 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr12_+_8666126 | 4.22 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr2_+_234580499 | 4.22 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr19_+_852291 | 4.21 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr3_-_148939835 | 4.19 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr2_+_234580525 | 4.14 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr12_-_123187890 | 4.11 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr11_-_102668879 | 4.09 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr6_-_49712123 | 4.04 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr12_-_123201337 | 4.04 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr6_+_31895467 | 4.03 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr6_-_49712147 | 4.01 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_-_88244010 | 3.98 |
ENST00000302219.6
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr11_-_116662593 | 3.98 |
ENST00000227665.4
|
APOA5
|
apolipoprotein A-V |
| chr2_-_21266935 | 3.98 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr7_+_137761167 | 3.97 |
ENST00000432161.1
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr3_+_186330712 | 3.96 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr4_+_74606223 | 3.95 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr7_+_137761199 | 3.87 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr19_+_840963 | 3.83 |
ENST00000234347.5
|
PRTN3
|
proteinase 3 |
| chr3_-_148939598 | 3.83 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr2_+_128177458 | 3.83 |
ENST00000409048.1
ENST00000422777.3 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr3_+_186435065 | 3.80 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr18_-_61311485 | 3.77 |
ENST00000436264.1
ENST00000356424.6 ENST00000341074.5 |
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr11_+_60050026 | 3.76 |
ENST00000395016.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr20_+_56136136 | 3.76 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr16_-_55866997 | 3.73 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr7_-_100239132 | 3.70 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr2_+_143635222 | 3.70 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr2_+_143635067 | 3.67 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr12_+_20963647 | 3.56 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr12_+_20963632 | 3.56 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr8_+_110656344 | 3.54 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
| chr18_+_61554932 | 3.51 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr1_+_186265399 | 3.49 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr12_-_53074182 | 3.46 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr10_+_74653330 | 3.41 |
ENST00000334011.5
|
OIT3
|
oncoprotein induced transcript 3 |
| chrX_+_12924732 | 3.38 |
ENST00000218032.6
ENST00000311912.5 |
TLR8
|
toll-like receptor 8 |
| chr19_+_6887571 | 3.35 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr2_+_234545092 | 3.31 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr19_-_6670128 | 3.31 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr1_+_196621156 | 3.29 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr4_-_88244049 | 3.24 |
ENST00000328546.4
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr10_+_114133773 | 3.20 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr4_-_185747188 | 3.19 |
ENST00000507295.1
ENST00000504900.1 ENST00000281455.2 ENST00000454703.2 |
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr10_+_26727125 | 3.14 |
ENST00000376236.4
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr11_-_10590238 | 3.08 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr3_+_157154578 | 3.07 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr22_+_35776828 | 3.04 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr3_-_46249878 | 3.04 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr6_+_30457244 | 3.01 |
ENST00000376630.4
|
HLA-E
|
major histocompatibility complex, class I, E |
| chr12_+_100897130 | 3.01 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr10_-_70092671 | 2.98 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr2_+_228678550 | 2.96 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr1_+_196857144 | 2.84 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr21_-_46348694 | 2.82 |
ENST00000355153.4
ENST00000397850.2 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr10_-_70092635 | 2.81 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr1_+_196621002 | 2.80 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr22_+_23010756 | 2.78 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr11_+_77774897 | 2.72 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr6_+_127898312 | 2.70 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr2_-_85895295 | 2.69 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr6_-_49712091 | 2.66 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr10_-_96829246 | 2.65 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chrX_-_131353461 | 2.63 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr5_-_100238918 | 2.63 |
ENST00000451528.2
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr8_+_18248755 | 2.61 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr19_+_55385682 | 2.60 |
ENST00000391726.3
|
FCAR
|
Fc fragment of IgA, receptor for |
| chr21_-_46348626 | 2.60 |
ENST00000517563.1
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_+_117297007 | 2.57 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr17_-_72619869 | 2.56 |
ENST00000392619.1
ENST00000426295.2 |
CD300E
|
CD300e molecule |
| chr17_-_34308524 | 2.56 |
ENST00000293275.3
|
CCL16
|
chemokine (C-C motif) ligand 16 |
| chr16_-_11692320 | 2.56 |
ENST00000571627.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr2_+_102608306 | 2.54 |
ENST00000332549.3
|
IL1R2
|
interleukin 1 receptor, type II |
| chr8_+_74271144 | 2.54 |
ENST00000519134.1
ENST00000518355.1 |
RP11-434I12.3
|
RP11-434I12.3 |
| chr12_-_8693469 | 2.53 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr2_-_31637560 | 2.53 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr2_-_21266816 | 2.50 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr1_-_203198790 | 2.47 |
ENST00000367229.1
ENST00000255427.3 ENST00000535569.1 |
CHIT1
|
chitinase 1 (chitotriosidase) |
| chr7_-_15601595 | 2.46 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr8_+_18248786 | 2.45 |
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr2_+_89999259 | 2.45 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr10_+_26727333 | 2.43 |
ENST00000356785.4
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr6_-_49712072 | 2.41 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr17_+_27369918 | 2.35 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr11_+_67777751 | 2.32 |
ENST00000316367.6
ENST00000007633.8 ENST00000342456.6 |
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr12_-_10251576 | 2.32 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr2_+_96331830 | 2.30 |
ENST00000425887.2
|
AC008268.1
|
AC008268.1 |
| chr20_-_1600642 | 2.30 |
ENST00000381603.3
ENST00000381605.4 ENST00000279477.7 ENST00000568365.1 ENST00000564763.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr22_-_30901637 | 2.29 |
ENST00000381982.3
ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4
|
SEC14-like 4 (S. cerevisiae) |
| chrX_-_131623874 | 2.29 |
ENST00000436215.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr6_+_106534192 | 2.29 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr14_+_21387491 | 2.28 |
ENST00000258817.2
|
RP11-84C10.2
|
RP11-84C10.2 |
| chr11_-_58980342 | 2.28 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr14_+_21423611 | 2.26 |
ENST00000304625.2
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr12_-_10251603 | 2.26 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr14_-_70263979 | 2.24 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr11_-_104905840 | 2.22 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr2_+_234826016 | 2.22 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr1_-_206945830 | 2.22 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr3_+_171844762 | 2.21 |
ENST00000443501.1
|
FNDC3B
|
fibronectin type III domain containing 3B |
| chr20_+_9966728 | 2.20 |
ENST00000449270.1
|
RP5-839B4.8
|
RP5-839B4.8 |
| chr3_+_108541608 | 2.20 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr1_+_111415757 | 2.19 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr10_-_45474237 | 2.19 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr14_+_95078714 | 2.18 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr17_-_34417479 | 2.16 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr8_-_126963387 | 2.15 |
ENST00000522865.1
ENST00000517869.1 |
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr3_+_186692745 | 2.14 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chrX_-_131623982 | 2.13 |
ENST00000370844.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr3_+_186358148 | 2.13 |
ENST00000382134.3
ENST00000265029.3 |
FETUB
|
fetuin B |
| chr3_-_49726104 | 2.09 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr14_-_106494587 | 2.09 |
ENST00000390597.2
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr3_-_151047327 | 2.08 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr3_-_194072019 | 2.07 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr2_-_89545079 | 2.05 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr11_+_60048053 | 2.04 |
ENST00000337908.4
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chrX_-_102348017 | 2.02 |
ENST00000425644.1
ENST00000395065.3 ENST00000425463.2 |
NXF3
|
nuclear RNA export factor 3 |
| chr1_-_89531041 | 2.00 |
ENST00000370473.4
|
GBP1
|
guanylate binding protein 1, interferon-inducible |
| chr17_-_73840415 | 2.00 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr13_+_78109884 | 1.99 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr2_+_234545148 | 1.99 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr13_+_78109804 | 1.98 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr4_-_70725856 | 1.97 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr11_+_7597639 | 1.97 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr19_-_40971643 | 1.97 |
ENST00000595483.1
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr11_+_67806467 | 1.96 |
ENST00000265686.3
ENST00000524598.1 ENST00000529657.1 |
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 |
| chr6_-_26199471 | 1.94 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr19_+_55417530 | 1.94 |
ENST00000350790.5
ENST00000338835.5 ENST00000357397.5 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr1_+_209941942 | 1.94 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr11_+_60048004 | 1.93 |
ENST00000532114.1
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr14_-_106573756 | 1.90 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr4_-_48136217 | 1.89 |
ENST00000264316.4
|
TXK
|
TXK tyrosine kinase |
| chr1_-_92952433 | 1.89 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr19_-_54876558 | 1.89 |
ENST00000391742.2
ENST00000434277.2 |
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr2_+_103035102 | 1.89 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr11_+_60048129 | 1.87 |
ENST00000355131.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr19_+_56652556 | 1.84 |
ENST00000337080.3
|
ZNF444
|
zinc finger protein 444 |
| chr5_-_180229833 | 1.83 |
ENST00000307826.4
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr12_+_21168630 | 1.82 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_-_49726486 | 1.82 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr17_+_34391625 | 1.81 |
ENST00000004921.3
|
CCL18
|
chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) |
| chr1_+_101185290 | 1.80 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr17_-_73840614 | 1.79 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr3_-_197300194 | 1.78 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr3_+_186358200 | 1.75 |
ENST00000382136.3
|
FETUB
|
fetuin B |
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 3.9 | 11.8 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 3.3 | 10.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 2.9 | 14.6 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 2.6 | 10.3 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 2.5 | 9.8 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 2.3 | 6.8 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 2.1 | 6.2 | GO:1904640 | response to methionine(GO:1904640) |
| 2.0 | 8.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 1.9 | 13.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 1.8 | 7.4 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.7 | 18.3 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 1.4 | 5.8 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.4 | 5.7 | GO:0035425 | autocrine signaling(GO:0035425) |
| 1.1 | 28.0 | GO:0008228 | opsonization(GO:0008228) |
| 1.1 | 6.5 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 1.1 | 26.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.0 | 3.0 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 1.0 | 3.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.9 | 3.8 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.9 | 3.7 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.9 | 16.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.9 | 5.3 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.9 | 8.5 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.8 | 7.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.8 | 5.0 | GO:0098759 | interleukin-8-mediated signaling pathway(GO:0038112) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.8 | 7.4 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.8 | 6.5 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.8 | 4.0 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.8 | 4.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.8 | 3.8 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.8 | 2.3 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.8 | 3.0 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.8 | 3.0 | GO:0038185 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.7 | 2.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.7 | 10.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.7 | 4.2 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.7 | 1.4 | GO:0090265 | positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.7 | 4.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.7 | 9.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.7 | 3.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.7 | 2.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 2.6 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.6 | 2.5 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.6 | 2.5 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.6 | 1.9 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.6 | 1.9 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.6 | 24.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.6 | 42.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.6 | 5.9 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.5 | 7.0 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.5 | 2.4 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.5 | 3.3 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.5 | 5.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.5 | 1.8 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.5 | 2.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 1.8 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.4 | 4.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 1.6 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.4 | 5.6 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.4 | 1.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.4 | 1.2 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.4 | 2.6 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.4 | 1.1 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.4 | 1.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 2.3 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.3 | 2.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 8.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.3 | 2.9 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 2.2 | GO:2000503 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 2.5 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.3 | 5.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 0.9 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.3 | 1.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 1.1 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 1.4 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) |
| 0.3 | 1.9 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 0.8 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.3 | 5.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 0.8 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.3 | 0.8 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.2 | 0.5 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.2 | 8.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 2.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 2.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 3.2 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 1.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 2.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.7 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.2 | 2.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 1.7 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.2 | 5.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 8.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 5.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 1.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 3.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 1.6 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.2 | 1.1 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.2 | 1.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 | 1.8 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 7.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.3 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.2 | 0.8 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.2 | 8.7 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 1.4 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 1.1 | GO:2000391 | positive regulation of neutrophil extravasation(GO:2000391) positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.2 | 1.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.9 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.7 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 17.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 1.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 13.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.2 | 3.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 4.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 1.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.2 | 0.3 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 0.5 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.2 | 0.5 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.2 | 1.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 1.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 8.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.5 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 1.3 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.9 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 17.7 | GO:2000257 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.1 | 3.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.7 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.4 | GO:0018969 | thiocyanate metabolic process(GO:0018969) |
| 0.1 | 4.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.5 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 2.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 3.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 2.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 4.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 1.2 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0070101 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 5.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 2.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 1.4 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 1.4 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 1.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 1.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 3.7 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 2.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.7 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 1.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.9 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 2.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.1 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.1 | 1.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 2.6 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.3 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 1.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 2.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.0 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.4 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 2.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.9 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 1.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 3.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.6 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.6 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 2.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 1.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.9 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 1.7 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.1 | 0.7 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.5 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 2.2 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 9.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.6 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 37.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.1 | 0.7 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 1.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.9 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 1.6 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.7 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 2.6 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 5.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0014894 | response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.2 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 1.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.0 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.8 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 1.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 2.7 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0071034 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.8 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 3.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 1.5 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.9 | GO:0070670 | response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 1.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 7.6 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 1.3 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.3 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 1.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 1.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 1.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 2.0 | 18.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.2 | 16.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.0 | 107.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.9 | 20.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.9 | 8.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.7 | 5.2 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.7 | 5.4 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.6 | 1.8 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.6 | 5.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 24.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 5.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 1.6 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.4 | 3.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 2.0 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 3.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.6 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.3 | 71.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 1.9 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.3 | 9.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 1.6 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.3 | 30.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 1.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 2.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 4.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.7 | GO:0097486 | alveolar lamellar body(GO:0097208) multivesicular body lumen(GO:0097486) |
| 0.2 | 0.8 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 1.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 1.2 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 3.6 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.2 | 0.6 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 29.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 1.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 3.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 5.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.1 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.1 | 1.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 6.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.9 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 17.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.2 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 2.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 1.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 4.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.1 | GO:0031975 | organelle envelope(GO:0031967) envelope(GO:0031975) |
| 0.0 | 4.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 15.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 7.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 5.3 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 39.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 3.3 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 3.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 2.9 | 11.8 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.9 | 17.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 2.8 | 13.8 | GO:0033265 | choline binding(GO:0033265) |
| 1.7 | 15.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.7 | 10.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.6 | 7.8 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 1.4 | 8.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.3 | 10.5 | GO:0035473 | lipase binding(GO:0035473) |
| 1.3 | 5.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.3 | 8.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 1.3 | 3.8 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.2 | 7.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.0 | 4.0 | GO:0019862 | IgA binding(GO:0019862) |
| 1.0 | 3.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.9 | 4.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.9 | 3.7 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.9 | 8.5 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.8 | 5.0 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.8 | 3.3 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.8 | 2.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.8 | 3.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.8 | 3.0 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.7 | 3.7 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.7 | 38.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.7 | 8.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.6 | 5.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.6 | 2.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.6 | 22.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.5 | 2.7 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 2.5 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.4 | 1.8 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.4 | 5.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.4 | 2.5 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.4 | 20.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 1.6 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.4 | 2.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.4 | 6.7 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 1.4 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.3 | 2.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 4.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 2.9 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 5.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 0.9 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.3 | 2.5 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.3 | 1.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 4.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 7.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 5.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 0.8 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.2 | 2.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 27.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 9.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 1.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 2.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 0.8 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 1.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 4.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 2.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 6.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 7.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 1.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 66.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 0.7 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 3.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 0.9 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.2 | 1.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 0.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 2.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 4.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 1.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 3.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 28.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.2 | 0.9 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 2.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 6.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.7 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 2.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0036393 | thiocyanate peroxidase activity(GO:0036393) |
| 0.1 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 2.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 9.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.9 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.1 | 1.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 5.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 12.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 2.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.5 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 2.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 2.1 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 2.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 3.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 4.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 2.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 3.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 9.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0097617 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.0 | 2.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.7 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 12.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 41.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 16.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 9.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 5.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 3.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 10.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 10.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 7.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 4.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 14.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 16.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 5.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 27.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 7.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 4.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 8.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 26.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 6.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 2.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 3.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 4.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 13.0 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 5.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 47.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.3 | 24.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.2 | 16.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 22.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 20.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.6 | 9.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.5 | 7.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 22.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.5 | 26.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.5 | 14.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 5.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 7.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.4 | 8.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 8.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 4.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 7.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 20.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 5.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 4.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 8.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 3.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 6.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 6.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 5.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 3.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 7.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 10.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 3.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 9.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 4.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.9 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 3.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 2.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 3.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 3.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 3.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 10.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.1 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 1.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 2.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 2.3 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 3.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.2 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 4.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 4.0 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |