Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for E2F8
Z-value: 0.24
Transcription factors associated with E2F8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F8
|
ENSG00000129173.8 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F8 | hg19_v2_chr11_-_19263145_19263176 | 0.67 | 2.4e-05 | Click! |
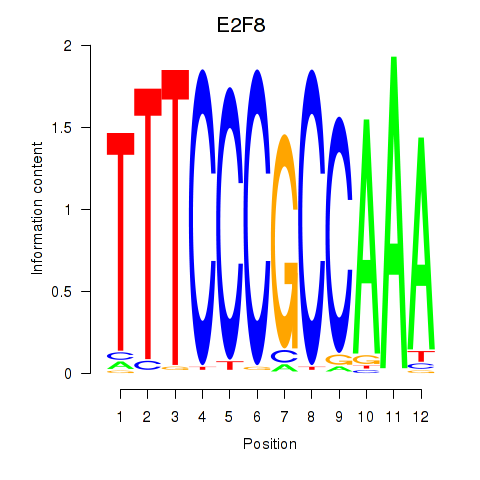
Activity profile of E2F8 motif
Sorted Z-values of E2F8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_24346218 | 3.54 |
ENST00000436622.1
ENST00000313213.4 |
PFN4
|
profilin family, member 4 |
| chr1_+_152850787 | 3.19 |
ENST00000368765.3
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein |
| chr2_+_79252804 | 2.87 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr2_-_198650037 | 2.84 |
ENST00000392296.4
|
BOLL
|
boule-like RNA-binding protein |
| chr13_-_19755975 | 2.83 |
ENST00000400113.3
|
TUBA3C
|
tubulin, alpha 3c |
| chr2_+_79252822 | 2.81 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr2_-_152589670 | 2.80 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr2_-_130956006 | 2.59 |
ENST00000312988.7
|
TUBA3E
|
tubulin, alpha 3e |
| chr2_+_132233664 | 2.39 |
ENST00000321253.6
|
TUBA3D
|
tubulin, alpha 3d |
| chr16_-_34740829 | 2.31 |
ENST00000562591.1
|
RP11-80F22.15
|
RP11-80F22.15 |
| chr19_+_49891475 | 2.28 |
ENST00000447857.3
|
CCDC155
|
coiled-coil domain containing 155 |
| chr2_+_79252834 | 2.25 |
ENST00000409471.1
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr1_-_150693318 | 2.09 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr2_-_27362263 | 2.06 |
ENST00000432962.1
ENST00000335524.3 |
C2orf53
|
chromosome 2 open reading frame 53 |
| chr8_+_17434689 | 1.97 |
ENST00000398074.3
|
PDGFRL
|
platelet-derived growth factor receptor-like |
| chr12_+_48499883 | 1.93 |
ENST00000546755.1
ENST00000549366.1 ENST00000552792.1 |
PFKM
|
phosphofructokinase, muscle |
| chr18_+_71983048 | 1.89 |
ENST00000579455.1
|
C18orf63
|
chromosome 18 open reading frame 63 |
| chr22_+_19467261 | 1.72 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr9_-_7961080 | 1.67 |
ENST00000435444.1
|
RP11-29B9.2
|
RP11-29B9.2 |
| chr10_-_131762105 | 1.64 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr1_-_150693305 | 1.63 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chrX_-_48216101 | 1.63 |
ENST00000298396.2
ENST00000376893.3 |
SSX3
|
synovial sarcoma, X breakpoint 3 |
| chr19_+_41103063 | 1.60 |
ENST00000308370.7
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr6_-_166401402 | 1.59 |
ENST00000581850.1
ENST00000444465.1 |
LINC00473
|
long intergenic non-protein coding RNA 473 |
| chr12_+_48499252 | 1.51 |
ENST00000549003.1
ENST00000550924.1 |
PFKM
|
phosphofructokinase, muscle |
| chr10_+_124670121 | 1.49 |
ENST00000368894.1
|
FAM24A
|
family with sequence similarity 24, member A |
| chr4_+_15341442 | 1.49 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chrX_-_52736211 | 1.43 |
ENST00000336777.5
ENST00000337502.5 |
SSX2
|
synovial sarcoma, X breakpoint 2 |
| chr21_+_19273574 | 1.39 |
ENST00000400128.1
|
CHODL
|
chondrolectin |
| chr22_-_29457832 | 1.39 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr2_+_74011316 | 1.37 |
ENST00000409561.1
|
C2orf78
|
chromosome 2 open reading frame 78 |
| chr3_-_107596910 | 1.33 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr15_-_89456630 | 1.32 |
ENST00000268150.8
|
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chrX_+_105445717 | 1.29 |
ENST00000372552.1
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr15_-_89456593 | 1.29 |
ENST00000558029.1
ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chrX_+_52240504 | 1.24 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr19_+_49891533 | 1.23 |
ENST00000598730.1
ENST00000594905.1 ENST00000595828.1 ENST00000594043.1 |
CCDC155
|
coiled-coil domain containing 155 |
| chrX_-_135338503 | 1.23 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr6_-_28973037 | 1.23 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr2_-_113993020 | 1.22 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr1_-_17307173 | 1.21 |
ENST00000438542.1
ENST00000375535.3 |
MFAP2
|
microfibrillar-associated protein 2 |
| chrX_+_130192318 | 1.19 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr9_-_96717654 | 1.17 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr19_-_35719609 | 1.14 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr10_+_118083919 | 1.13 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr17_-_37123646 | 1.13 |
ENST00000378079.2
|
FBXO47
|
F-box protein 47 |
| chr11_-_19262486 | 1.10 |
ENST00000250024.4
|
E2F8
|
E2F transcription factor 8 |
| chrX_+_130192216 | 1.09 |
ENST00000276211.5
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr18_+_21718924 | 1.08 |
ENST00000399496.3
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr3_-_46875313 | 1.08 |
ENST00000447340.1
|
PRSS42
|
protease, serine, 42 |
| chr6_-_166401442 | 1.07 |
ENST00000584179.1
|
LINC00473
|
long intergenic non-protein coding RNA 473 |
| chr2_-_177502659 | 1.01 |
ENST00000295549.4
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr6_+_88117683 | 1.01 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr5_-_145483932 | 1.00 |
ENST00000311450.4
|
PLAC8L1
|
PLAC8-like 1 |
| chr11_-_110968081 | 1.00 |
ENST00000603154.1
|
RP11-89C3.4
|
RP11-89C3.4 |
| chr9_-_74675521 | 1.00 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr4_+_152330409 | 0.99 |
ENST00000513086.1
|
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr2_+_238395879 | 0.99 |
ENST00000445024.2
ENST00000338530.4 ENST00000409373.1 |
MLPH
|
melanophilin |
| chr11_-_19263145 | 0.98 |
ENST00000532666.1
ENST00000527884.1 |
E2F8
|
E2F transcription factor 8 |
| chr8_+_106331769 | 0.97 |
ENST00000520492.1
|
ZFPM2
|
zinc finger protein, FOG family member 2 |
| chr17_-_27949911 | 0.97 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr3_+_44803322 | 0.97 |
ENST00000481166.2
|
KIF15
|
kinesin family member 15 |
| chr12_+_3068957 | 0.96 |
ENST00000543035.1
|
TEAD4
|
TEA domain family member 4 |
| chr3_+_127317705 | 0.95 |
ENST00000480910.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr4_+_128702969 | 0.94 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chrX_-_48271344 | 0.94 |
ENST00000376884.2
ENST00000396928.1 |
SSX4B
|
synovial sarcoma, X breakpoint 4B |
| chr4_-_152329987 | 0.93 |
ENST00000508847.1
|
RP11-610P16.1
|
RP11-610P16.1 |
| chr4_-_141348763 | 0.93 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr3_+_44803209 | 0.92 |
ENST00000326047.4
|
KIF15
|
kinesin family member 15 |
| chr6_-_57086200 | 0.90 |
ENST00000468148.1
|
RAB23
|
RAB23, member RAS oncogene family |
| chr11_-_107729287 | 0.88 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_-_8815215 | 0.88 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr15_+_84115868 | 0.87 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr14_+_102027688 | 0.86 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr6_-_50016364 | 0.85 |
ENST00000322246.4
|
DEFB112
|
defensin, beta 112 |
| chr2_-_208994548 | 0.84 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr3_+_69985792 | 0.84 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr12_-_8815299 | 0.84 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr7_-_100026280 | 0.82 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr7_+_75932863 | 0.81 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr14_-_67878917 | 0.81 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr10_+_106028923 | 0.80 |
ENST00000338595.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr11_+_9595180 | 0.79 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr17_+_42733803 | 0.78 |
ENST00000409122.2
|
C17orf104
|
chromosome 17 open reading frame 104 |
| chr13_+_19756173 | 0.78 |
ENST00000382988.2
|
RP11-408E5.4
|
RP11-408E5.4 |
| chr19_+_51153045 | 0.77 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr18_+_21719018 | 0.77 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr8_+_22411931 | 0.77 |
ENST00000523402.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr17_-_15165854 | 0.76 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
| chr12_+_3069037 | 0.76 |
ENST00000397122.2
|
TEAD4
|
TEA domain family member 4 |
| chr10_+_32856764 | 0.76 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr3_+_44754126 | 0.76 |
ENST00000449836.1
ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr10_+_115312766 | 0.75 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr9_+_124088860 | 0.74 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr3_-_123168551 | 0.73 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr12_-_48499591 | 0.73 |
ENST00000551330.1
ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr7_-_21985489 | 0.73 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr16_+_11439286 | 0.73 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr14_-_61747949 | 0.72 |
ENST00000355702.2
|
TMEM30B
|
transmembrane protein 30B |
| chr3_-_146187088 | 0.72 |
ENST00000497985.1
|
PLSCR2
|
phospholipid scramblase 2 |
| chr19_-_19144243 | 0.72 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr16_-_31147020 | 0.72 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr9_-_138591341 | 0.71 |
ENST00000298466.5
ENST00000425225.1 |
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| chr18_+_29078131 | 0.71 |
ENST00000585206.1
|
DSG2
|
desmoglein 2 |
| chr3_+_14058794 | 0.70 |
ENST00000424053.1
ENST00000528067.1 ENST00000429201.1 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr15_-_88247083 | 0.69 |
ENST00000560439.1
|
RP11-648K4.2
|
RP11-648K4.2 |
| chr19_+_6740888 | 0.69 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr15_-_40074996 | 0.69 |
ENST00000350221.3
|
FSIP1
|
fibrous sheath interacting protein 1 |
| chr14_-_61190754 | 0.69 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr20_+_42295745 | 0.68 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr14_-_55658252 | 0.67 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr4_-_141348999 | 0.67 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr14_-_55658323 | 0.67 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr8_-_120868078 | 0.67 |
ENST00000313655.4
|
DSCC1
|
DNA replication and sister chromatid cohesion 1 |
| chr4_-_135248604 | 0.67 |
ENST00000515491.1
ENST00000504728.1 ENST00000506638.1 |
RP11-400D2.2
|
RP11-400D2.2 |
| chr11_-_30105890 | 0.66 |
ENST00000527819.1
|
RP11-624D11.2
|
RP11-624D11.2 |
| chr11_-_107729504 | 0.66 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_-_57113281 | 0.64 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr11_+_134201911 | 0.64 |
ENST00000389881.3
|
GLB1L2
|
galactosidase, beta 1-like 2 |
| chr17_+_47075023 | 0.63 |
ENST00000431824.2
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr17_-_15165825 | 0.63 |
ENST00000426385.3
|
PMP22
|
peripheral myelin protein 22 |
| chr9_+_139690784 | 0.63 |
ENST00000338005.6
|
KIAA1984
|
coiled-coil domain containing 183 |
| chr16_-_31146961 | 0.62 |
ENST00000567531.1
|
PRSS8
|
protease, serine, 8 |
| chr3_+_127317066 | 0.62 |
ENST00000265056.7
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr19_-_49828438 | 0.61 |
ENST00000454748.3
ENST00000598828.1 ENST00000335875.4 |
SLC6A16
|
solute carrier family 6, member 16 |
| chr11_-_71639613 | 0.61 |
ENST00000528184.1
ENST00000528511.2 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr11_-_83435965 | 0.61 |
ENST00000434967.1
ENST00000530800.1 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr22_+_24105241 | 0.61 |
ENST00000402217.3
|
C22orf15
|
chromosome 22 open reading frame 15 |
| chr8_+_27632083 | 0.60 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr3_+_140770183 | 0.60 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr17_+_39975455 | 0.60 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr14_+_24837226 | 0.59 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr4_-_141348789 | 0.59 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr6_+_160693591 | 0.59 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr1_-_146989697 | 0.58 |
ENST00000437831.1
|
LINC00624
|
long intergenic non-protein coding RNA 624 |
| chr9_-_23821273 | 0.58 |
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_-_39928106 | 0.58 |
ENST00000540235.1
|
JUP
|
junction plakoglobin |
| chr3_-_25824925 | 0.57 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr10_-_46167722 | 0.57 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr10_-_17659234 | 0.56 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr1_-_36916011 | 0.56 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr6_+_135502466 | 0.56 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_+_55096489 | 0.55 |
ENST00000504461.1
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr1_+_43637996 | 0.55 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr19_-_17559376 | 0.55 |
ENST00000341130.5
|
TMEM221
|
transmembrane protein 221 |
| chr17_+_7184986 | 0.54 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr12_-_102455846 | 0.54 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr12_-_102455902 | 0.54 |
ENST00000240079.6
|
CCDC53
|
coiled-coil domain containing 53 |
| chr1_-_32827682 | 0.54 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229, member A |
| chr17_+_42733730 | 0.54 |
ENST00000359945.3
ENST00000425535.1 |
C17orf104
|
chromosome 17 open reading frame 104 |
| chr17_-_56769382 | 0.53 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr1_-_27709816 | 0.53 |
ENST00000374030.1
|
CD164L2
|
CD164 sialomucin-like 2 |
| chr10_-_17659357 | 0.53 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr19_-_56704421 | 0.52 |
ENST00000358992.3
|
ZSCAN5B
|
zinc finger and SCAN domain containing 5B |
| chr17_+_39975544 | 0.52 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr18_-_19994830 | 0.52 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chrX_+_24073048 | 0.52 |
ENST00000423068.1
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr14_-_23624511 | 0.51 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr3_-_185542761 | 0.50 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_+_12203100 | 0.50 |
ENST00000596883.1
|
ZNF788
|
zinc finger family member 788 |
| chrX_-_130423386 | 0.50 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr1_-_36916066 | 0.49 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr6_+_24357131 | 0.49 |
ENST00000274766.1
|
KAAG1
|
kidney associated antigen 1 |
| chr19_+_12203069 | 0.49 |
ENST00000430298.2
ENST00000339302.4 |
ZNF788
ZNF788
|
zinc finger family member 788 Zinc finger protein 788 |
| chr3_-_151176497 | 0.48 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr12_+_75760714 | 0.48 |
ENST00000547144.1
|
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr3_-_79816965 | 0.48 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr10_+_35416223 | 0.47 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr1_+_33283246 | 0.47 |
ENST00000526230.1
ENST00000531256.1 ENST00000482212.1 |
S100PBP
|
S100P binding protein |
| chr12_-_66275350 | 0.47 |
ENST00000536648.1
|
RP11-366L20.2
|
Uncharacterized protein |
| chr12_-_6798025 | 0.47 |
ENST00000542351.1
ENST00000538829.1 |
ZNF384
|
zinc finger protein 384 |
| chr19_-_47291843 | 0.46 |
ENST00000542575.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr11_+_101981423 | 0.46 |
ENST00000531439.1
|
YAP1
|
Yes-associated protein 1 |
| chr11_+_125496124 | 0.45 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr1_-_158656488 | 0.45 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr1_+_33283043 | 0.44 |
ENST00000373476.1
ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP
|
S100P binding protein |
| chr17_-_46623441 | 0.43 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr11_-_73882249 | 0.42 |
ENST00000535954.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr17_+_46125707 | 0.41 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr6_-_84937314 | 0.41 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr22_+_40742512 | 0.41 |
ENST00000454266.2
ENST00000342312.6 |
ADSL
|
adenylosuccinate lyase |
| chr17_+_46125685 | 0.41 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr7_-_21985656 | 0.40 |
ENST00000406877.3
|
CDCA7L
|
cell division cycle associated 7-like |
| chr2_-_37193606 | 0.40 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr7_+_128379449 | 0.40 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr1_-_55230165 | 0.40 |
ENST00000371279.3
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr3_-_185542817 | 0.40 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr1_-_27709793 | 0.40 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr10_+_35416090 | 0.39 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr6_+_117002339 | 0.39 |
ENST00000413340.1
ENST00000368564.1 ENST00000356348.1 |
KPNA5
|
karyopherin alpha 5 (importin alpha 6) |
| chr6_-_2962331 | 0.39 |
ENST00000380524.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr7_+_128379346 | 0.39 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr2_-_230787879 | 0.38 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr17_+_46126135 | 0.38 |
ENST00000361665.3
ENST00000585062.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr2_+_233385173 | 0.38 |
ENST00000449534.2
|
PRSS56
|
protease, serine, 56 |
| chr22_+_40742497 | 0.38 |
ENST00000216194.7
|
ADSL
|
adenylosuccinate lyase |
| chr15_+_40731920 | 0.38 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr18_-_12656715 | 0.37 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr17_-_9479128 | 0.37 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr14_+_36295504 | 0.37 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.2 | 3.7 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.7 | 2.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.6 | 3.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.6 | 1.7 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.6 | 2.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 1.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.4 | 3.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.3 | 4.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 1.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 1.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 | 1.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 0.8 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.2 | 1.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.2 | 0.5 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 | 0.7 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.2 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.2 | 0.8 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 0.5 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 4.0 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.2 | 2.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 2.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.4 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.7 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 0.1 | 0.7 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 1.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.4 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.5 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 0.7 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.8 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 0.3 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 2.8 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.8 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.3 | GO:0018012 | N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.1 | 0.6 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 2.0 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.1 | 0.2 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 1.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.9 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.3 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.5 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 2.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.3 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.1 | 0.8 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.1 | 0.3 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.1 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.2 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 1.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.4 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.7 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 2.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 1.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 1.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.9 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.4 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.7 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 1.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 1.2 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.1 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 1.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.0 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.5 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 1.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.7 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 1.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.4 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.5 | 1.9 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.4 | 3.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 3.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 0.9 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 0.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.6 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.1 | 1.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 3.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 1.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 2.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.4 | 7.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.3 | 3.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.8 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.2 | 0.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.2 | 0.6 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.2 | 1.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 2.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.6 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.8 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.1 | 0.4 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 2.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 2.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.9 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 3.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.3 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.1 | 0.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.2 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 3.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 2.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 2.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 6.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 7.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 2.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 9.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 5.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |