Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for EGR3_EGR2
Z-value: 0.34
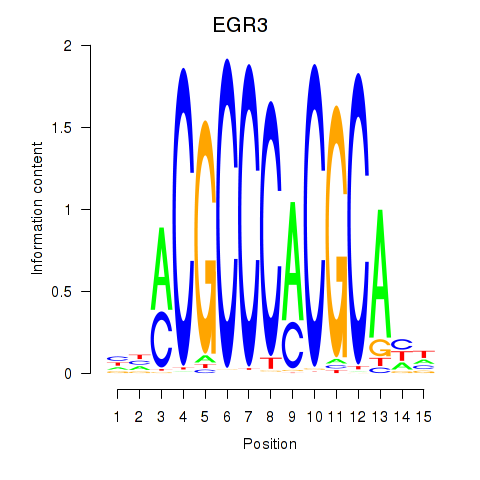
Transcription factors associated with EGR3_EGR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR3
|
ENSG00000179388.8 | early growth response 3 |
|
EGR2
|
ENSG00000122877.9 | early growth response 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR3 | hg19_v2_chr8_-_22550815_22550844 | 0.60 | 2.7e-04 | Click! |
| EGR2 | hg19_v2_chr10_-_64576105_64576133 | 0.12 | 5.1e-01 | Click! |
Activity profile of EGR3_EGR2 motif
Sorted Z-values of EGR3_EGR2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_84115868 | 5.27 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr11_+_124609823 | 3.97 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr11_+_124609742 | 3.90 |
ENST00000284292.6
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chrX_+_117957741 | 3.51 |
ENST00000310164.2
|
ZCCHC12
|
zinc finger, CCHC domain containing 12 |
| chr15_+_84116106 | 3.43 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr19_+_10400615 | 3.15 |
ENST00000221980.4
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin |
| chr16_+_25703274 | 3.07 |
ENST00000331351.5
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr11_-_18813110 | 2.86 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr19_+_49617581 | 2.71 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr4_-_90758227 | 2.63 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_37782955 | 2.61 |
ENST00000580825.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr19_-_13617247 | 2.58 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr20_+_37353084 | 2.44 |
ENST00000217420.1
|
SLC32A1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr6_-_110500826 | 2.42 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr19_+_49617609 | 2.40 |
ENST00000221459.2
ENST00000486217.2 |
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr19_+_589893 | 2.37 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr4_+_150999418 | 2.36 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chrX_-_128788914 | 2.36 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr1_-_35395178 | 2.36 |
ENST00000373347.1
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr4_-_90758118 | 2.30 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_+_99282570 | 2.29 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr19_+_35634146 | 2.28 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr11_-_18765389 | 2.25 |
ENST00000477854.1
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr5_+_7396141 | 2.25 |
ENST00000338316.4
|
ADCY2
|
adenylate cyclase 2 (brain) |
| chr6_-_110500905 | 2.20 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr9_+_140772226 | 2.19 |
ENST00000277551.2
ENST00000371372.1 ENST00000277549.5 ENST00000371363.1 ENST00000371357.1 ENST00000371355.4 |
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr11_-_18813353 | 2.17 |
ENST00000358540.2
ENST00000396171.4 ENST00000396167.2 |
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chrX_-_47479246 | 2.17 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr2_+_220306745 | 2.11 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chrX_+_49126294 | 2.08 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr19_-_40724246 | 2.06 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr22_+_19702069 | 2.06 |
ENST00000412544.1
|
SEPT5
|
septin 5 |
| chr7_-_100493482 | 2.00 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr17_+_30593195 | 2.00 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr15_-_83316711 | 1.99 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr12_+_79258444 | 1.98 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr19_-_51472031 | 1.98 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr20_+_44035200 | 1.97 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr12_-_65515334 | 1.96 |
ENST00000286574.4
|
WIF1
|
WNT inhibitory factor 1 |
| chr8_+_73449625 | 1.90 |
ENST00000523207.1
|
KCNB2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr9_+_34958254 | 1.90 |
ENST00000242315.3
|
KIAA1045
|
KIAA1045 |
| chr15_-_71146480 | 1.90 |
ENST00000299213.8
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr21_+_34398153 | 1.89 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr2_-_61108449 | 1.85 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr19_-_51472222 | 1.81 |
ENST00000376851.3
|
KLK6
|
kallikrein-related peptidase 6 |
| chr19_+_54412517 | 1.78 |
ENST00000391767.1
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7 |
| chr19_+_55795493 | 1.78 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr12_-_49393092 | 1.77 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr10_+_105036909 | 1.77 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr17_-_42992856 | 1.75 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr3_-_149688655 | 1.74 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr20_+_17207636 | 1.74 |
ENST00000262545.2
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr11_+_66059339 | 1.73 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr7_-_100253993 | 1.73 |
ENST00000461605.1
ENST00000160382.5 |
ACTL6B
|
actin-like 6B |
| chr19_-_38714847 | 1.72 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr1_+_23695680 | 1.71 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr6_+_72596604 | 1.68 |
ENST00000348717.5
ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr17_+_37783197 | 1.67 |
ENST00000582680.1
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr18_+_33877654 | 1.66 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr17_-_36760865 | 1.65 |
ENST00000584266.1
|
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr5_+_137774706 | 1.65 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr17_+_37783170 | 1.64 |
ENST00000254079.4
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr6_-_110501126 | 1.62 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chr7_+_94537542 | 1.62 |
ENST00000433881.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr14_-_77737543 | 1.56 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr16_-_850723 | 1.55 |
ENST00000248150.4
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr6_-_110501200 | 1.55 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr7_+_24323782 | 1.55 |
ENST00000242152.2
ENST00000407573.1 |
NPY
|
neuropeptide Y |
| chrX_+_130192216 | 1.55 |
ENST00000276211.5
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr14_-_23822080 | 1.54 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr16_-_62070305 | 1.54 |
ENST00000584337.1
|
CDH8
|
cadherin 8, type 2 |
| chr4_+_158142750 | 1.54 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr18_-_70534745 | 1.54 |
ENST00000583169.1
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr1_+_6845384 | 1.54 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr8_-_57233103 | 1.53 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr12_+_70760056 | 1.53 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr3_+_123813509 | 1.52 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr8_+_22019168 | 1.52 |
ENST00000318561.3
ENST00000521315.1 ENST00000437090.2 ENST00000520605.1 ENST00000522109.1 ENST00000524255.1 ENST00000523296.1 ENST00000518615.1 |
SFTPC
|
surfactant protein C |
| chr19_-_36523709 | 1.50 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr4_-_36246060 | 1.49 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_+_95401851 | 1.49 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chrX_-_152939133 | 1.49 |
ENST00000370150.1
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr20_+_17207665 | 1.48 |
ENST00000536609.1
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr12_+_79258547 | 1.48 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr15_+_43809797 | 1.48 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr11_+_66045634 | 1.46 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr2_+_210636697 | 1.46 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr22_-_38380543 | 1.45 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr19_-_46145696 | 1.42 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr14_+_100150622 | 1.42 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr17_-_77179487 | 1.42 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr2_-_42721110 | 1.41 |
ENST00000394973.4
ENST00000306078.1 |
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr19_-_13617037 | 1.40 |
ENST00000360228.5
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr1_-_38512450 | 1.40 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr15_+_51633826 | 1.38 |
ENST00000335449.6
|
GLDN
|
gliomedin |
| chr8_-_75233563 | 1.38 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr10_-_135150367 | 1.37 |
ENST00000368555.3
ENST00000252939.4 ENST00000368558.1 ENST00000368556.2 |
CALY
|
calcyon neuron-specific vesicular protein |
| chr1_-_160040038 | 1.36 |
ENST00000368089.3
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr21_+_22370717 | 1.36 |
ENST00000284894.7
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr21_+_22370608 | 1.36 |
ENST00000400546.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr16_-_31021717 | 1.35 |
ENST00000565419.1
|
STX1B
|
syntaxin 1B |
| chr6_-_84419101 | 1.34 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr7_-_44365020 | 1.33 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr3_-_49907323 | 1.33 |
ENST00000296471.7
ENST00000488336.1 ENST00000467248.1 ENST00000466940.1 ENST00000463537.1 ENST00000480398.2 |
CAMKV
|
CaM kinase-like vesicle-associated |
| chr8_-_144242020 | 1.33 |
ENST00000414417.2
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr18_-_70210764 | 1.32 |
ENST00000585159.1
ENST00000584764.1 |
CBLN2
|
cerebellin 2 precursor |
| chr22_+_19701985 | 1.31 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr3_-_149688896 | 1.30 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr20_+_44034676 | 1.30 |
ENST00000372723.3
ENST00000372722.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr14_+_29234870 | 1.28 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr9_+_140033862 | 1.28 |
ENST00000350902.5
ENST00000371550.4 ENST00000371546.4 ENST00000371555.4 ENST00000371553.3 ENST00000371559.4 ENST00000371560.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr12_+_3600356 | 1.27 |
ENST00000382622.3
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr7_+_95401877 | 1.26 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr3_-_149688971 | 1.26 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr15_-_71146460 | 1.26 |
ENST00000344870.4
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chrX_-_152939252 | 1.25 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr3_+_35681081 | 1.23 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr13_+_26042960 | 1.22 |
ENST00000255283.8
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
| chr19_-_41196458 | 1.22 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr12_-_57634475 | 1.22 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr5_+_161274685 | 1.20 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr16_+_5009250 | 1.20 |
ENST00000587469.1
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae) |
| chr3_-_149688502 | 1.20 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chrX_-_49056635 | 1.19 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr1_+_237205476 | 1.18 |
ENST00000366574.2
|
RYR2
|
ryanodine receptor 2 (cardiac) |
| chr15_+_79575073 | 1.17 |
ENST00000421388.2
|
ANKRD34C
|
ankyrin repeat domain 34C |
| chr8_-_110986918 | 1.17 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr5_+_161274940 | 1.17 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr8_-_132052458 | 1.17 |
ENST00000377928.3
|
ADCY8
|
adenylate cyclase 8 (brain) |
| chr16_+_77822427 | 1.16 |
ENST00000302536.2
|
VAT1L
|
vesicle amine transport 1-like |
| chr14_+_77648167 | 1.16 |
ENST00000554346.1
ENST00000298351.4 |
TMEM63C
|
transmembrane protein 63C |
| chr19_-_36523529 | 1.15 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr15_+_32322685 | 1.14 |
ENST00000454250.3
ENST00000306901.3 |
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| chr3_-_52002194 | 1.13 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr16_-_31021921 | 1.13 |
ENST00000215095.5
|
STX1B
|
syntaxin 1B |
| chr2_-_219433014 | 1.12 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr5_+_161275320 | 1.12 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr17_+_20059358 | 1.11 |
ENST00000536879.1
ENST00000395522.2 ENST00000395525.3 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr17_+_20059302 | 1.11 |
ENST00000395530.2
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr10_-_43762329 | 1.09 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr11_-_125366089 | 1.09 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr14_+_94640633 | 1.09 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr16_-_49891694 | 1.07 |
ENST00000562520.1
|
ZNF423
|
zinc finger protein 423 |
| chr3_+_71803201 | 1.07 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr12_+_51984657 | 1.07 |
ENST00000550891.1
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chrX_-_53350522 | 1.06 |
ENST00000396435.3
ENST00000375368.5 |
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr15_-_27184664 | 1.06 |
ENST00000541819.2
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr16_+_5008290 | 1.04 |
ENST00000251170.7
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae) |
| chr20_-_41818373 | 1.04 |
ENST00000373187.1
ENST00000356100.2 ENST00000373184.1 ENST00000373190.1 |
PTPRT
|
protein tyrosine phosphatase, receptor type, T |
| chr15_-_83316254 | 1.03 |
ENST00000567678.1
ENST00000450751.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr19_+_34112850 | 1.03 |
ENST00000591231.1
ENST00000434302.1 ENST00000438847.3 |
CHST8
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8 |
| chr7_+_103969104 | 1.03 |
ENST00000424859.1
ENST00000535008.1 ENST00000401970.2 ENST00000543266.1 |
LHFPL3
|
lipoma HMGIC fusion partner-like 3 |
| chr16_+_2198604 | 1.02 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr5_-_176037105 | 1.02 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr2_+_220495800 | 1.02 |
ENST00000413743.1
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr7_+_153749732 | 1.01 |
ENST00000377770.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chrX_-_140271249 | 1.01 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1 |
| chr5_+_149569520 | 1.00 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr7_-_108096822 | 1.00 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr3_+_238427 | 1.00 |
ENST00000397491.2
|
CHL1
|
cell adhesion molecule L1-like |
| chrX_+_70364667 | 0.99 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr15_-_80695917 | 0.99 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr6_+_72596406 | 0.98 |
ENST00000491071.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr1_-_37499726 | 0.98 |
ENST00000373091.3
ENST00000373093.4 |
GRIK3
|
glutamate receptor, ionotropic, kainate 3 |
| chr1_-_23694794 | 0.98 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr4_+_114038137 | 0.97 |
ENST00000508613.1
|
ANK2
|
ankyrin 2, neuronal |
| chr19_-_41196534 | 0.97 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr12_-_55375622 | 0.97 |
ENST00000316577.8
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr4_+_83351715 | 0.96 |
ENST00000273920.3
|
ENOPH1
|
enolase-phosphatase 1 |
| chr20_-_40247002 | 0.96 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr14_+_94640671 | 0.96 |
ENST00000328839.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr8_-_9760839 | 0.96 |
ENST00000519461.1
ENST00000517675.1 |
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chrX_+_130192318 | 0.96 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr7_+_86274145 | 0.95 |
ENST00000439827.1
ENST00000394720.2 ENST00000421579.1 |
GRM3
|
glutamate receptor, metabotropic 3 |
| chr7_-_108096765 | 0.94 |
ENST00000379024.4
ENST00000351718.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr16_+_23847267 | 0.94 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr19_-_12945362 | 0.94 |
ENST00000590404.1
ENST00000592204.1 |
RTBDN
|
retbindin |
| chr11_+_67183141 | 0.94 |
ENST00000531040.1
ENST00000307823.3 ENST00000423745.2 |
CARNS1
|
carnosine synthase 1 |
| chrX_-_48814810 | 0.94 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr13_-_37494365 | 0.92 |
ENST00000350148.5
|
SMAD9
|
SMAD family member 9 |
| chr8_-_57232656 | 0.92 |
ENST00000396721.2
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr19_+_30017406 | 0.92 |
ENST00000335523.7
|
VSTM2B
|
V-set and transmembrane domain containing 2B |
| chr10_-_75255724 | 0.92 |
ENST00000342558.3
ENST00000360663.5 ENST00000394829.2 ENST00000394828.2 ENST00000394822.2 |
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr20_-_21494654 | 0.91 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr7_-_100493744 | 0.91 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr1_-_205313304 | 0.90 |
ENST00000539253.1
ENST00000607826.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr17_-_79008373 | 0.90 |
ENST00000577066.1
ENST00000573167.1 |
BAIAP2-AS1
|
BAIAP2 antisense RNA 1 (head to head) |
| chr14_+_65170820 | 0.89 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr6_-_40555176 | 0.89 |
ENST00000338305.6
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr3_+_167453026 | 0.89 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr19_-_3801789 | 0.89 |
ENST00000590849.1
ENST00000395045.2 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr3_-_50541028 | 0.89 |
ENST00000266039.3
ENST00000435965.1 ENST00000395083.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr20_+_53092123 | 0.89 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr11_+_64685026 | 0.89 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chrX_+_71996972 | 0.88 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr13_+_88324870 | 0.88 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr13_-_38443860 | 0.88 |
ENST00000426868.2
ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr9_+_129986734 | 0.88 |
ENST00000444677.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr11_-_125366018 | 0.88 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr15_+_32322709 | 0.87 |
ENST00000455693.2
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR3_EGR2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.9 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.9 | 2.8 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.8 | 0.8 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.8 | 2.4 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.7 | 2.9 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.7 | 10.0 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.7 | 4.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.6 | 3.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.6 | 3.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.6 | 2.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.6 | 5.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.5 | 5.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.5 | 2.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.5 | 1.9 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.5 | 1.9 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.4 | 1.3 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.4 | 1.3 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.4 | 2.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.4 | 5.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 1.2 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.4 | 1.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 1.5 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.4 | 1.4 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.3 | 1.9 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 0.9 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.3 | 1.5 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 1.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 0.9 | GO:0060929 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.3 | 11.0 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.3 | 2.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.3 | 1.1 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.3 | 2.6 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.3 | 5.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 2.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 3.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.2 | 1.8 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.2 | 0.7 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 1.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 0.9 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 2.5 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.8 | GO:1903060 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.2 | 2.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 2.4 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.2 | 0.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 1.4 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.2 | 1.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.2 | 0.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 2.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.6 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.2 | 2.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 0.7 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.2 | 3.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 1.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 2.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 0.9 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 3.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 1.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 1.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 2.6 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.2 | 0.8 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 2.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 0.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 1.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 1.5 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.2 | 1.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 7.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 1.0 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.4 | GO:0061193 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 1.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.4 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.1 | 0.7 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 1.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.1 | 0.5 | GO:0071469 | sensory perception of touch(GO:0050975) detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 2.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.9 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.8 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.4 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.1 | 0.5 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 2.7 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.6 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 1.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 1.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.6 | GO:0043474 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 1.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.2 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 0.9 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.5 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 2.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.7 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 1.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 2.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 0.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 1.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 2.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 1.0 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.5 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.1 | 0.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.3 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 3.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 1.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 1.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.3 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.4 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.5 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 1.4 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 1.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.4 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.6 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.4 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.1 | 1.3 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.9 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 5.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 2.5 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 0.4 | GO:0070458 | establishment of blood-nerve barrier(GO:0008065) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.3 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 2.4 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.3 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 1.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.4 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.4 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.3 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.2 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.1 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.3 | GO:2000672 | response to sorbitol(GO:0072708) cellular response to sorbitol(GO:0072709) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.2 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.9 | GO:0021756 | striatum development(GO:0021756) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 1.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0061346 | chemoattraction of serotonergic neuron axon(GO:0036517) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 1.1 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 1.0 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 2.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 1.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.6 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.3 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.0 | 0.7 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 1.3 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 7.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.5 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.3 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.2 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 2.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.4 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 1.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 6.7 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.4 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 1.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.0 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.9 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.4 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 1.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.9 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 1.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.0 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.4 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.4 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.7 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.1 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.1 | GO:0060084 | micturition(GO:0060073) synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.4 | GO:0034349 | glial cell apoptotic process(GO:0034349) |
| 0.0 | 0.2 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.3 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.6 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 1.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:1902462 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 1.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.0 | GO:0090194 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) negative regulation of glomerulus development(GO:0090194) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.0 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) kidney mesenchymal cell proliferation(GO:0072135) metanephric mesenchymal cell proliferation involved in metanephros development(GO:0072136) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.5 | 9.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 7.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 2.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 6.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 1.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.4 | 3.5 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.3 | 5.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 1.1 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 0.8 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 1.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 1.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 5.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 2.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 3.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.6 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 0.6 | GO:1990742 | microvesicle(GO:1990742) |
| 0.2 | 5.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 0.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 17.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 8.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 4.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 1.5 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 1.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.9 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 2.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 3.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 2.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 2.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 2.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 5.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.4 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 2.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 9.2 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.1 | 0.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 11.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 5.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 8.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.1 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 1.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0035859 | Seh1-associated complex(GO:0035859) Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 9.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 1.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 1.0 | 4.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.6 | 2.9 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.5 | 1.9 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.5 | 4.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 2.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 2.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.4 | 1.3 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.4 | 2.4 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.4 | 5.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.4 | 1.1 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.3 | 2.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.3 | 1.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.3 | 1.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.3 | 3.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 8.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 2.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.3 | 2.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 2.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 8.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 2.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 0.5 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.2 | 1.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 1.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 0.6 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 4.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 0.9 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.2 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 1.7 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 1.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.9 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.1 | 0.4 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.3 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 3.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.0 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.6 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 3.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 3.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 2.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 6.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 7.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 4.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 10.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.3 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.7 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 2.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.6 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 2.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 2.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.9 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 2.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.2 | GO:0070546 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 1.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0052840 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.1 | 2.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 3.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.1 | 0.3 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 2.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.8 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 3.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 2.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 2.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.1 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 2.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 1.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 1.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.7 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 2.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 5.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 1.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.7 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 6.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 2.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 1.3 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 10.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 3.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 4.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 6.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 3.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 9.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 5.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 6.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.9 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.1 | 4.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 3.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 6.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 18.7 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.1 | 1.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 4.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 2.9 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 1.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |