Project
Illumina Body Map 2, young vs old
Navigation
Downloads
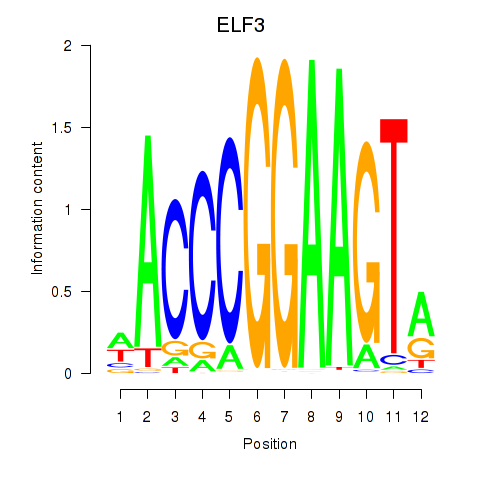
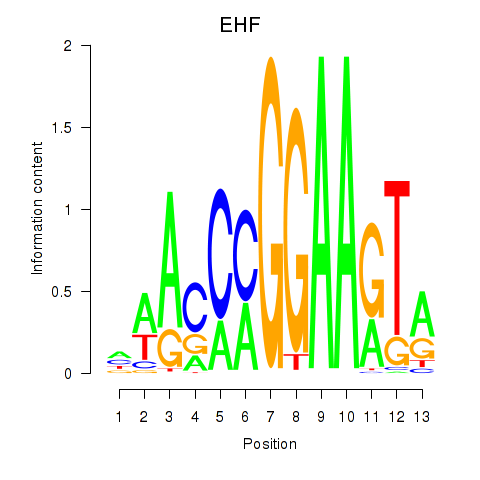
Results for ELF3_EHF
Z-value: 0.87
Transcription factors associated with ELF3_EHF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ELF3
|
ENSG00000163435.11 | E74 like ETS transcription factor 3 |
|
EHF
|
ENSG00000135373.8 | ETS homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EHF | hg19_v2_chr11_+_34645791_34645836 | 0.65 | 6.5e-05 | Click! |
| ELF3 | hg19_v2_chr1_+_201979743_201979772 | 0.56 | 9.6e-04 | Click! |
Activity profile of ELF3_EHF motif
Sorted Z-values of ELF3_EHF motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ELF3_EHF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.0 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.1 | 1.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.0 | 6.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.0 | 3.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 1.0 | 7.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.0 | 1.9 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.8 | 2.3 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.8 | 2.3 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.7 | 2.9 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.7 | 2.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.7 | 2.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.7 | 5.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.7 | 2.0 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.6 | 2.6 | GO:0061642 | chemoattraction of serotonergic neuron axon(GO:0036517) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.6 | 2.5 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.5 | 3.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.5 | 1.6 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.5 | 2.6 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.5 | 7.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.5 | 8.6 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.5 | 2.5 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.5 | 2.5 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.5 | 1.5 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.5 | 0.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.5 | 2.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 | 2.3 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.5 | 1.4 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) positive regulation of response to alcohol(GO:1901421) |
| 0.5 | 1.8 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.4 | 1.7 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.4 | 2.4 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.4 | 2.0 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.4 | 1.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.4 | 2.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.4 | 1.9 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.4 | 4.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.4 | 9.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 0.7 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.4 | 0.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.3 | 1.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.3 | 8.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 1.0 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.3 | 5.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 2.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 2.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.3 | 13.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.3 | 2.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 1.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.3 | 1.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.3 | 0.9 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 0.8 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.3 | 1.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.3 | 0.8 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.3 | 1.3 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.3 | 1.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.3 | 0.8 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.3 | 1.8 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.2 | 0.7 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 1.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 0.7 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.2 | 1.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.2 | 0.7 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 1.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 0.9 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.2 | 3.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 1.3 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 3.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 2.8 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.2 | 0.6 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 1.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.6 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.2 | 0.8 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.2 | 4.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 1.6 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 1.4 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 0.6 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.2 | 0.6 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.2 | 1.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 3.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.2 | 0.5 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.2 | 1.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 2.9 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.2 | 0.7 | GO:2000619 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.2 | 0.5 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.2 | 3.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 4.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 0.5 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.2 | 0.5 | GO:0061566 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.2 | 0.5 | GO:0031622 | positive regulation of fever generation(GO:0031622) |
| 0.2 | 1.0 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 0.8 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 | 2.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 1.9 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 1.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.2 | 0.3 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.2 | 0.9 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 4.8 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.2 | 1.4 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 0.8 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 0.6 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 1.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 2.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 1.6 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 1.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.7 | GO:2001053 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.4 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.1 | 2.9 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 1.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 3.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 1.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.8 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 1.4 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.4 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 0.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 1.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.8 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.4 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.5 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.4 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.6 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 1.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 9.5 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.1 | 0.4 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 2.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.8 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 0.6 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.6 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.4 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 1.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 2.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.7 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 1.4 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.1 | 1.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.9 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.8 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 1.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.3 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.5 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) negative regulation of sperm motility(GO:1901318) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.1 | 5.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.7 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0039020 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric nephron tubule development(GO:0039020) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.1 | 0.4 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 0.3 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.3 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 0.9 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.3 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 2.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 4.0 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 1.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.3 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.2 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 1.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.6 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.1 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.8 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 | 1.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 1.8 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 1.5 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.4 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.1 | 0.3 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.1 | 1.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.6 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 1.6 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.6 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 1.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.9 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 2.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.3 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 2.0 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 2.0 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 2.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.9 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.1 | 1.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.5 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.2 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.1 | 0.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 1.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 3.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.4 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.5 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 1.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 4.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.2 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.1 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 0.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.3 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.5 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 3.5 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 1.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.5 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 0.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.2 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.6 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.5 | GO:0007369 | gastrulation(GO:0007369) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 4.3 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 1.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 7.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.4 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 1.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.7 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.1 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 1.1 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.7 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.2 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.1 | 1.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.7 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 0.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.3 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.6 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.7 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.5 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 1.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 1.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 6.2 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 1.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 2.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.8 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 1.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.4 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 1.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 3.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 1.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.5 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 3.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 2.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.5 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 2.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 4.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.2 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.6 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 1.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.9 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.2 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0009635 | response to herbicide(GO:0009635) response to vitamin B1(GO:0010266) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.3 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 1.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 3.0 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 1.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.7 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.2 | GO:0072350 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) tricarboxylic acid metabolic process(GO:0072350) |
| 0.0 | 1.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.9 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.5 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 1.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 1.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 1.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.5 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 2.1 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 1.1 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.5 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.4 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.9 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.2 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.5 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 1.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.0 | 1.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.4 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0021683 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 1.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.3 | GO:0061572 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 1.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 3.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.7 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 1.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.8 | 2.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.7 | 4.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.6 | 1.9 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.4 | 6.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.4 | 2.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 3.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.4 | 4.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 12.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 2.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.3 | 0.8 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.2 | 1.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 0.9 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 2.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 4.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 1.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 3.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 19.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 2.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 0.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 2.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.7 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.9 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 2.5 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 1.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 3.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 4.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.8 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.6 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 8.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.5 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 2.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 0.4 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 0.1 | 1.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 0.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 4.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 2.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.5 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.9 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 1.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 9.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 1.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 12.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 7.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.6 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 6.2 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 4.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.8 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 13.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 8.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0061673 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 11.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 5.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.6 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 2.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.8 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 1.4 | 4.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.9 | 3.7 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.9 | 3.5 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.7 | 4.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.7 | 4.8 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.7 | 9.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.6 | 2.6 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.6 | 6.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 1.8 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.6 | 3.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.6 | 2.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.5 | 1.6 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.5 | 4.9 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 2.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.5 | 1.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.4 | 2.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 10.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.0 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 0.8 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.3 | 1.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.3 | 0.8 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.3 | 1.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 1.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 1.6 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.3 | 6.0 | GO:0030215 | semaphorin receptor activity(GO:0017154) semaphorin receptor binding(GO:0030215) |
| 0.3 | 1.5 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.3 | 0.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 1.0 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.2 | 0.7 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.2 | 8.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 9.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 2.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.5 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 0.8 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 0.6 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.2 | 3.0 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 2.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.6 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 1.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 0.6 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 0.9 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.2 | 1.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.2 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 1.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 1.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 0.9 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 1.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 0.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 3.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.6 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.2 | 1.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 4.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 1.7 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 2.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.2 | 0.8 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 0.5 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.1 | 3.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.4 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 18.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 1.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 6.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 2.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 2.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.4 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.6 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.3 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 0.7 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 2.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.6 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 3.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.4 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 2.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.5 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 1.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 2.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.6 | GO:0019871 | potassium channel inhibitor activity(GO:0019870) sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 2.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 3.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.8 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.9 | GO:0070739 | NEDD8 transferase activity(GO:0019788) protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 0.6 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 2.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 5.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 2.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.5 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 1.0 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 1.7 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 3.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 6.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.5 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.2 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 2.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 1.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 2.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.2 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.1 | 0.5 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.0 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 5.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 7.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 3.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 1.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.6 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 1.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 1.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 2.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 1.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 1.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 2.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 7.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 1.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 3.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 9.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 9.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 7.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 3.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 9.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 7.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.3 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | REACTOME REGULATION OF MITOTIC CELL CYCLE | Genes involved in Regulation of mitotic cell cycle |
| 0.3 | 15.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 12.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 4.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 6.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 8.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 6.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 1.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 4.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 4.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 4.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 4.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.8 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 2.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 2.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 16.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 2.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 3.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 1.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |