Project
Illumina Body Map 2, young vs old
Navigation
Downloads
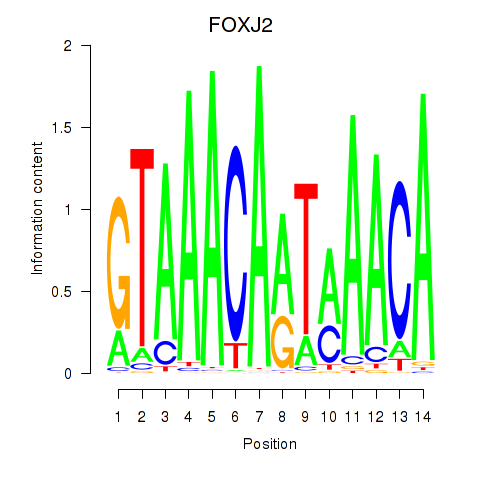
Results for FOXJ2
Z-value: 0.05
Transcription factors associated with FOXJ2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXJ2
|
ENSG00000065970.4 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ2 | hg19_v2_chr12_+_8185288_8185339 | 0.66 | 4.2e-05 | Click! |
Activity profile of FOXJ2 motif
Sorted Z-values of FOXJ2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_134029937 | 1.92 |
ENST00000518108.1
|
TG
|
thyroglobulin |
| chr19_+_11658655 | 1.87 |
ENST00000588935.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr7_+_73242069 | 1.58 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr12_-_91574142 | 1.57 |
ENST00000547937.1
|
DCN
|
decorin |
| chr15_-_56757329 | 1.55 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr19_-_55677920 | 1.38 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr8_+_104383728 | 1.37 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr8_+_104383759 | 1.33 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr4_+_68424434 | 1.30 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr13_-_38172863 | 1.28 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr12_-_53074182 | 1.25 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr10_-_105992059 | 1.24 |
ENST00000369720.1
ENST00000369719.1 ENST00000357060.3 ENST00000428666.1 ENST00000278064.2 |
WDR96
|
WD repeat domain 96 |
| chr7_-_80548667 | 1.22 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_-_182921119 | 1.22 |
ENST00000423786.1
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr20_-_42355629 | 1.20 |
ENST00000373003.1
|
GTSF1L
|
gametocyte specific factor 1-like |
| chr7_-_80548493 | 1.17 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr11_-_5537920 | 1.11 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr1_-_72566613 | 1.10 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr5_-_138780159 | 1.06 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr2_-_24346218 | 1.01 |
ENST00000436622.1
ENST00000313213.4 |
PFN4
|
profilin family, member 4 |
| chr1_+_73771844 | 1.01 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr14_-_36990354 | 0.96 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chrX_-_6453159 | 0.91 |
ENST00000381089.3
ENST00000398729.1 |
VCX3A
|
variable charge, X-linked 3A |
| chr11_-_125550726 | 0.90 |
ENST00000315608.3
ENST00000530048.1 |
ACRV1
|
acrosomal vesicle protein 1 |
| chr7_+_72742178 | 0.88 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr5_-_110062349 | 0.88 |
ENST00000511883.2
ENST00000455884.2 |
TMEM232
|
transmembrane protein 232 |
| chrX_+_114524275 | 0.85 |
ENST00000371921.1
ENST00000451986.2 ENST00000371920.3 |
LUZP4
|
leucine zipper protein 4 |
| chr6_-_10747802 | 0.83 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr19_-_14196574 | 0.81 |
ENST00000548523.1
ENST00000343945.5 |
C19orf67
|
chromosome 19 open reading frame 67 |
| chrX_+_79591003 | 0.80 |
ENST00000538312.1
|
FAM46D
|
family with sequence similarity 46, member D |
| chr12_-_8815215 | 0.78 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr8_-_95274536 | 0.78 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr3_-_100551141 | 0.77 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr12_+_24376201 | 0.76 |
ENST00000540733.1
ENST00000539583.1 |
RP11-778H2.1
|
RP11-778H2.1 |
| chr14_-_54418598 | 0.76 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr1_+_197881592 | 0.76 |
ENST00000367391.1
ENST00000367390.3 |
LHX9
|
LIM homeobox 9 |
| chr1_+_45212074 | 0.75 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr4_-_90756769 | 0.74 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_+_56989609 | 0.73 |
ENST00000601875.1
|
ZNF667-AS1
|
ZNF667 antisense RNA 1 (head to head) |
| chrX_-_8139308 | 0.72 |
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2 |
| chr1_+_91966384 | 0.72 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chr19_+_48248779 | 0.72 |
ENST00000246802.5
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2 |
| chr8_+_103991013 | 0.71 |
ENST00000517983.1
|
KB-1507C5.4
|
KB-1507C5.4 |
| chr5_-_159739483 | 0.70 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr4_-_90757364 | 0.69 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_162602244 | 0.69 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr4_-_159956333 | 0.67 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr5_-_96209315 | 0.67 |
ENST00000504056.1
|
CTD-2260A17.2
|
Uncharacterized protein |
| chr11_-_125550764 | 0.67 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1 |
| chrX_+_7810303 | 0.67 |
ENST00000381059.3
ENST00000341408.4 |
VCX
|
variable charge, X-linked |
| chr3_+_132036243 | 0.65 |
ENST00000475741.1
ENST00000351273.7 |
ACPP
|
acid phosphatase, prostate |
| chrX_+_22056165 | 0.65 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chrX_-_139047669 | 0.64 |
ENST00000370540.1
|
CXorf66
|
chromosome X open reading frame 66 |
| chr10_-_23528745 | 0.64 |
ENST00000376501.5
|
C10orf115
|
chromosome 10 open reading frame 115 |
| chr14_+_37126765 | 0.63 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr15_+_74165945 | 0.63 |
ENST00000535547.2
ENST00000300504.2 ENST00000562056.1 |
TBC1D21
|
TBC1 domain family, member 21 |
| chr2_+_132160448 | 0.62 |
ENST00000437751.1
|
AC073869.19
|
long intergenic non-protein coding RNA 1120 |
| chr12_-_8815404 | 0.62 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr20_-_45530365 | 0.62 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr13_+_43355732 | 0.61 |
ENST00000313851.1
|
FAM216B
|
family with sequence similarity 216, member B |
| chr1_+_150122034 | 0.60 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chrX_+_99899180 | 0.59 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr10_-_111713633 | 0.58 |
ENST00000538668.1
ENST00000369657.1 ENST00000369655.1 |
RP11-451M19.3
|
Uncharacterized protein |
| chrX_+_8432871 | 0.58 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr3_+_157154578 | 0.58 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr6_+_31082603 | 0.56 |
ENST00000259881.9
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr1_+_239882842 | 0.56 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr5_-_110062384 | 0.55 |
ENST00000429839.2
|
TMEM232
|
transmembrane protein 232 |
| chr4_-_123542224 | 0.54 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr12_-_8815299 | 0.54 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr19_+_56989485 | 0.53 |
ENST00000585445.1
ENST00000586091.1 ENST00000594783.1 ENST00000592146.1 ENST00000588158.1 ENST00000299997.4 ENST00000591797.1 |
ZNF667-AS1
|
ZNF667 antisense RNA 1 (head to head) |
| chr19_+_49866331 | 0.53 |
ENST00000597873.1
|
DKKL1
|
dickkopf-like 1 |
| chr12_-_102874330 | 0.53 |
ENST00000307046.8
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr7_+_142334156 | 0.52 |
ENST00000390394.3
|
TRBV20-1
|
T cell receptor beta variable 20-1 |
| chr1_-_197115818 | 0.51 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr14_-_52066664 | 0.51 |
ENST00000556617.1
|
FRMD6-AS2
|
FRMD6 antisense RNA 2 |
| chr1_+_244624678 | 0.51 |
ENST00000366534.4
ENST00000366533.4 ENST00000428042.1 ENST00000366531.3 |
C1orf101
|
chromosome 1 open reading frame 101 |
| chr6_-_27858570 | 0.50 |
ENST00000359303.2
|
HIST1H3J
|
histone cluster 1, H3j |
| chr8_-_10336885 | 0.50 |
ENST00000520494.1
|
RP11-981G7.3
|
RP11-981G7.3 |
| chr10_-_49459800 | 0.49 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr2_+_210518057 | 0.49 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_+_43855560 | 0.49 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr14_+_38264418 | 0.48 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr12_+_108523133 | 0.46 |
ENST00000547525.1
|
WSCD2
|
WSC domain containing 2 |
| chrX_+_56644819 | 0.46 |
ENST00000446028.1
|
RP11-431N15.2
|
RP11-431N15.2 |
| chr21_+_47013566 | 0.46 |
ENST00000441095.1
ENST00000424569.1 |
AL133493.2
|
AL133493.2 |
| chr2_-_39414848 | 0.45 |
ENST00000451199.1
|
CDKL4
|
cyclin-dependent kinase-like 4 |
| chr15_-_55881135 | 0.45 |
ENST00000302000.6
|
PYGO1
|
pygopus family PHD finger 1 |
| chr12_-_7596735 | 0.43 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr14_-_52066633 | 0.42 |
ENST00000557625.1
|
FRMD6-AS2
|
FRMD6 antisense RNA 2 |
| chr1_-_16763685 | 0.42 |
ENST00000540400.1
|
SPATA21
|
spermatogenesis associated 21 |
| chr2_+_90121477 | 0.41 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr12_-_8815477 | 0.41 |
ENST00000433590.2
|
MFAP5
|
microfibrillar associated protein 5 |
| chr1_+_222988464 | 0.41 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr18_-_73967160 | 0.41 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr12_-_45444873 | 0.40 |
ENST00000332700.6
|
DBX2
|
developing brain homeobox 2 |
| chr8_-_16424871 | 0.39 |
ENST00000518026.1
|
MSR1
|
macrophage scavenger receptor 1 |
| chr12_+_1099675 | 0.39 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr10_+_50887683 | 0.39 |
ENST00000374113.3
ENST00000374111.3 ENST00000374112.3 ENST00000535836.1 |
C10orf53
|
chromosome 10 open reading frame 53 |
| chr21_-_27945562 | 0.38 |
ENST00000299340.4
ENST00000435845.2 |
CYYR1
|
cysteine/tyrosine-rich 1 |
| chrX_+_107683096 | 0.38 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr3_+_48413709 | 0.37 |
ENST00000296438.5
ENST00000436231.1 ENST00000445170.1 ENST00000415155.1 |
FBXW12
|
F-box and WD repeat domain containing 12 |
| chr19_-_55874611 | 0.36 |
ENST00000424985.3
|
FAM71E2
|
family with sequence similarity 71, member E2 |
| chr1_+_32739733 | 0.35 |
ENST00000333070.4
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr12_-_10251539 | 0.35 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr16_+_86609939 | 0.35 |
ENST00000593625.1
|
FOXL1
|
forkhead box L1 |
| chr1_+_149239529 | 0.34 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr6_-_52628271 | 0.32 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr1_+_43855545 | 0.32 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr4_+_147145709 | 0.32 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr2_-_239140276 | 0.32 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr9_-_130679257 | 0.31 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr2_+_120770581 | 0.31 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr10_+_95848824 | 0.30 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr8_-_33424636 | 0.30 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr20_+_23420322 | 0.30 |
ENST00000347397.1
|
CSTL1
|
cystatin-like 1 |
| chr12_-_10251603 | 0.30 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chrX_+_108780347 | 0.29 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chrX_-_107682702 | 0.28 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr2_+_90060377 | 0.28 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr19_-_54619006 | 0.28 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chrX_+_102024075 | 0.27 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr12_-_10251576 | 0.27 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr1_-_43855479 | 0.27 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr19_-_49865639 | 0.27 |
ENST00000593945.1
ENST00000601519.1 ENST00000539846.1 ENST00000596757.1 ENST00000311227.2 |
TEAD2
|
TEA domain family member 2 |
| chr1_+_160765947 | 0.27 |
ENST00000263285.6
ENST00000368039.2 |
LY9
|
lymphocyte antigen 9 |
| chrX_+_48687283 | 0.27 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr1_+_22778337 | 0.26 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr6_+_76599809 | 0.26 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr2_+_179318295 | 0.26 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr17_-_40215075 | 0.26 |
ENST00000436535.3
|
ZNF385C
|
zinc finger protein 385C |
| chr14_+_22591276 | 0.25 |
ENST00000390455.3
|
TRAV26-1
|
T cell receptor alpha variable 26-1 |
| chr15_+_64428529 | 0.25 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr3_+_26735991 | 0.25 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr3_+_132036207 | 0.25 |
ENST00000336375.5
ENST00000495911.1 |
ACPP
|
acid phosphatase, prostate |
| chr21_-_27945464 | 0.25 |
ENST00000400043.3
|
CYYR1
|
cysteine/tyrosine-rich 1 |
| chr2_-_89459813 | 0.25 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr4_+_26344754 | 0.25 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_-_130543958 | 0.24 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr11_+_6226782 | 0.24 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr13_+_28813645 | 0.24 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr15_-_34628951 | 0.23 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr15_+_66797455 | 0.23 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr18_+_32455201 | 0.23 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr12_-_120763739 | 0.23 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr2_-_85641162 | 0.22 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr17_+_73629500 | 0.22 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr8_+_30244580 | 0.22 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr4_-_68749745 | 0.21 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr8_+_101170257 | 0.20 |
ENST00000251809.3
|
SPAG1
|
sperm associated antigen 1 |
| chr2_-_239140011 | 0.20 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr1_-_156269428 | 0.20 |
ENST00000339922.3
|
VHLL
|
von Hippel-Lindau tumor suppressor-like |
| chr1_-_43855444 | 0.20 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr4_-_9390709 | 0.20 |
ENST00000508324.1
|
RP11-1396O13.13
|
Uncharacterized protein |
| chrX_+_11311533 | 0.20 |
ENST00000380714.3
ENST00000380712.3 ENST00000348912.4 |
AMELX
|
amelogenin, X-linked |
| chr3_+_127317705 | 0.20 |
ENST00000480910.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr6_+_31462658 | 0.19 |
ENST00000538442.1
|
MICB
|
MHC class I polypeptide-related sequence B |
| chr15_+_32933866 | 0.19 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr1_+_32739714 | 0.18 |
ENST00000461712.2
ENST00000373562.3 ENST00000477031.2 ENST00000373557.2 |
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr12_-_102874378 | 0.18 |
ENST00000456098.1
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr11_+_35201826 | 0.18 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chrX_+_55744166 | 0.18 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr19_+_47538560 | 0.18 |
ENST00000439365.2
ENST00000594670.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chrX_-_118699325 | 0.18 |
ENST00000320339.4
ENST00000371594.4 ENST00000536133.1 |
CXorf56
|
chromosome X open reading frame 56 |
| chr14_-_80782219 | 0.17 |
ENST00000553594.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr15_-_96590126 | 0.17 |
ENST00000561051.1
|
RP11-4G2.1
|
RP11-4G2.1 |
| chr17_-_2117600 | 0.17 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr2_+_162016827 | 0.17 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_66918558 | 0.17 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr8_+_101170134 | 0.16 |
ENST00000520643.1
|
SPAG1
|
sperm associated antigen 1 |
| chr1_+_160765919 | 0.16 |
ENST00000341032.4
ENST00000368041.2 ENST00000368040.1 |
LY9
|
lymphocyte antigen 9 |
| chr14_-_75735986 | 0.16 |
ENST00000553510.1
|
RP11-293M10.1
|
Uncharacterized protein |
| chr19_+_57752200 | 0.16 |
ENST00000414468.2
|
ZNF805
|
zinc finger protein 805 |
| chr19_+_54619125 | 0.16 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr7_+_23637763 | 0.16 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr9_-_21142144 | 0.16 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr16_-_90096309 | 0.15 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr2_+_210517895 | 0.15 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_+_160765860 | 0.15 |
ENST00000368037.5
|
LY9
|
lymphocyte antigen 9 |
| chr11_+_827248 | 0.15 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr9_-_98268883 | 0.15 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr19_+_55996316 | 0.14 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr12_-_102874416 | 0.14 |
ENST00000392904.1
ENST00000337514.6 |
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr1_+_107599267 | 0.14 |
ENST00000361318.5
ENST00000370078.1 |
PRMT6
|
protein arginine methyltransferase 6 |
| chr15_-_95676078 | 0.14 |
ENST00000554787.1
|
RP11-255M2.2
|
RP11-255M2.2 |
| chr19_+_57751973 | 0.14 |
ENST00000535550.1
|
ZNF805
|
zinc finger protein 805 |
| chr6_-_121655552 | 0.14 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr1_+_81106951 | 0.13 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr4_-_140005443 | 0.13 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr3_+_127317945 | 0.13 |
ENST00000472731.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr15_+_66797627 | 0.13 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr2_+_47454054 | 0.13 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr16_-_4817129 | 0.13 |
ENST00000545009.1
ENST00000219478.6 |
ZNF500
|
zinc finger protein 500 |
| chr10_+_28822636 | 0.12 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr3_-_49131788 | 0.12 |
ENST00000395443.2
ENST00000411682.1 |
QRICH1
|
glutamine-rich 1 |
| chr12_-_123728548 | 0.12 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr11_-_57417405 | 0.12 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr14_+_75761099 | 0.11 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr17_+_37617721 | 0.11 |
ENST00000584632.1
|
CDK12
|
cyclin-dependent kinase 12 |
| chr7_+_143792141 | 0.11 |
ENST00000408949.2
|
OR2A12
|
olfactory receptor, family 2, subfamily A, member 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXJ2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.3 | 2.7 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.3 | 1.4 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.9 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.2 | 2.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.2 | 1.3 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.2 | 0.8 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.8 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.9 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.2 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.8 | GO:0072097 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) deltoid tuberosity development(GO:0035993) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 1.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.8 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.7 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 1.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.6 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 1.9 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 2.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 1.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.2 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.1 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 2.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:1905068 | arterial endothelial cell fate commitment(GO:0060844) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.6 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.9 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 2.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 1.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.5 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.3 | GO:0043025 | somatodendritic compartment(GO:0036477) neuronal cell body(GO:0043025) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 0.9 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.2 | 1.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 1.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.7 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 2.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 1.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 3.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0070990 | U4 snRNA binding(GO:0030621) snRNP binding(GO:0070990) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |