Project
Illumina Body Map 2, young vs old
Navigation
Downloads
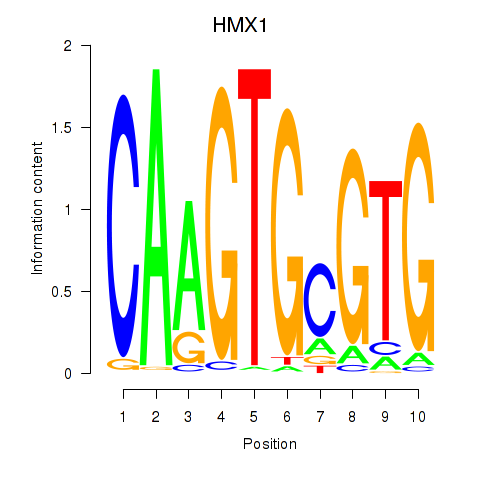
Results for HMX1
Z-value: 0.23
Transcription factors associated with HMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX1
|
ENSG00000215612.5 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX1 | hg19_v2_chr4_-_8873531_8873543 | 0.07 | 6.9e-01 | Click! |
Activity profile of HMX1 motif
Sorted Z-values of HMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_142326335 | 4.93 |
ENST00000390393.3
|
TRBV19
|
T cell receptor beta variable 19 |
| chr7_+_50344289 | 4.02 |
ENST00000413698.1
ENST00000359197.5 ENST00000331340.3 ENST00000357364.4 ENST00000343574.5 ENST00000349824.4 ENST00000346667.4 ENST00000440768.2 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr2_+_204801471 | 3.96 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr16_-_776431 | 3.05 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr11_-_64815333 | 2.90 |
ENST00000533842.1
ENST00000532802.1 ENST00000530139.1 ENST00000526516.1 |
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr1_-_167059830 | 2.87 |
ENST00000367868.3
|
GPA33
|
glycoprotein A33 (transmembrane) |
| chr6_-_31550192 | 2.59 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr12_-_55378470 | 2.57 |
ENST00000524668.1
ENST00000533607.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr16_-_86542652 | 2.54 |
ENST00000599749.1
|
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr16_+_28996364 | 2.53 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr7_-_16844611 | 2.48 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr1_-_25256368 | 2.45 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr13_+_43148281 | 2.39 |
ENST00000239849.6
ENST00000398795.2 ENST00000544862.1 |
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11 |
| chr11_-_128392085 | 2.39 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr20_+_30639991 | 2.30 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr20_+_30640004 | 2.24 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chr11_+_45944190 | 2.24 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr22_-_37640277 | 2.23 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chrX_+_13587712 | 2.19 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr16_-_86542455 | 2.18 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr4_-_109087445 | 2.15 |
ENST00000512172.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr1_-_207095212 | 2.13 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr3_+_47021168 | 2.13 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr15_+_81489213 | 1.94 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr22_-_37640456 | 1.94 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr4_-_109087906 | 1.92 |
ENST00000515500.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr4_-_109087872 | 1.91 |
ENST00000510624.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr17_-_72709050 | 1.89 |
ENST00000583937.1
ENST00000301573.9 ENST00000326165.6 ENST00000469092.1 |
CD300LF
|
CD300 molecule-like family member f |
| chr2_-_175462456 | 1.89 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chrX_+_131157322 | 1.87 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr14_-_25103472 | 1.86 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr4_-_2420335 | 1.82 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chrX_+_131157290 | 1.81 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr16_+_28996416 | 1.80 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr17_-_8868991 | 1.80 |
ENST00000447110.1
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr19_-_45579762 | 1.75 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr11_+_826136 | 1.73 |
ENST00000528315.1
ENST00000533803.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr6_-_32157947 | 1.72 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr17_-_7165662 | 1.71 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chrX_-_153599578 | 1.67 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr14_-_25103388 | 1.67 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr12_-_55378452 | 1.66 |
ENST00000449076.1
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr16_+_11439286 | 1.66 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr12_+_4382917 | 1.65 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr6_+_41606176 | 1.55 |
ENST00000441667.1
ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI
|
MyoD family inhibitor |
| chr19_+_6464243 | 1.52 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chrX_+_47444613 | 1.48 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr19_-_2308127 | 1.48 |
ENST00000404279.1
|
LINGO3
|
leucine rich repeat and Ig domain containing 3 |
| chr12_+_7060432 | 1.47 |
ENST00000318974.9
ENST00000456013.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_+_101546827 | 1.47 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr6_-_7911042 | 1.45 |
ENST00000379757.4
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr11_-_67275542 | 1.42 |
ENST00000531506.1
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr12_+_7060508 | 1.42 |
ENST00000541698.1
ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr6_-_42418999 | 1.41 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr12_+_7060414 | 1.41 |
ENST00000538715.1
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr17_+_74381343 | 1.39 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr2_-_175462934 | 1.39 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr16_-_67190152 | 1.37 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr15_-_38852251 | 1.37 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr11_-_417308 | 1.32 |
ENST00000397632.3
ENST00000382520.2 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr16_+_28996572 | 1.30 |
ENST00000360872.5
ENST00000566177.1 ENST00000354453.4 |
LAT
|
linker for activation of T cells |
| chr9_-_130331297 | 1.29 |
ENST00000373312.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr5_-_158636512 | 1.28 |
ENST00000424310.2
|
RNF145
|
ring finger protein 145 |
| chr14_-_55369525 | 1.26 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr12_+_57482665 | 1.23 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr16_+_50280020 | 1.22 |
ENST00000564965.1
|
ADCY7
|
adenylate cyclase 7 |
| chr19_-_46526304 | 1.22 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr2_+_203499901 | 1.20 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr2_-_219925189 | 1.20 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr1_-_207095324 | 1.19 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr2_+_10091783 | 1.15 |
ENST00000324883.5
|
GRHL1
|
grainyhead-like 1 (Drosophila) |
| chr7_+_130131907 | 1.15 |
ENST00000223215.4
ENST00000437945.1 |
MEST
|
mesoderm specific transcript |
| chr5_-_81046841 | 1.15 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr15_+_90611465 | 1.15 |
ENST00000559360.1
|
ZNF710
|
zinc finger protein 710 |
| chr17_-_79876010 | 1.14 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr13_+_42031679 | 1.14 |
ENST00000379359.3
|
RGCC
|
regulator of cell cycle |
| chr4_-_2420357 | 1.14 |
ENST00000511071.1
ENST00000509171.1 ENST00000290974.2 |
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr2_+_10091815 | 1.13 |
ENST00000324907.9
|
GRHL1
|
grainyhead-like 1 (Drosophila) |
| chr6_+_33043703 | 1.12 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr3_-_49066811 | 1.12 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr1_-_201476274 | 1.11 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr8_-_67525473 | 1.11 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr8_-_67525524 | 1.10 |
ENST00000517885.1
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr17_-_8151353 | 1.09 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr22_-_39268308 | 1.09 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr17_-_79212884 | 1.08 |
ENST00000300714.3
|
ENTHD2
|
ENTH domain containing 2 |
| chr2_+_30454390 | 1.08 |
ENST00000395323.3
ENST00000406087.1 ENST00000404397.1 |
LBH
|
limb bud and heart development |
| chr5_-_81046904 | 1.07 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr7_-_127672146 | 1.06 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr5_+_141016508 | 1.04 |
ENST00000444782.1
ENST00000521367.1 ENST00000297164.3 |
RELL2
|
RELT-like 2 |
| chr9_-_131940526 | 1.04 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr19_-_52227221 | 1.01 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr11_-_417388 | 1.01 |
ENST00000332725.3
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr1_+_156030937 | 1.00 |
ENST00000361084.5
|
RAB25
|
RAB25, member RAS oncogene family |
| chr17_+_74723031 | 0.99 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr19_+_1407517 | 0.99 |
ENST00000336761.6
ENST00000233078.4 |
DAZAP1
|
DAZ associated protein 1 |
| chr4_-_80993854 | 0.98 |
ENST00000346652.6
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr5_-_81046922 | 0.98 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr17_-_78450398 | 0.97 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr2_-_192016276 | 0.96 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr17_-_27278304 | 0.95 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr21_-_36259445 | 0.94 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr22_+_37447771 | 0.94 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr2_-_74776586 | 0.94 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr7_+_12726623 | 0.93 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr13_-_76111945 | 0.93 |
ENST00000355801.4
ENST00000406936.3 |
COMMD6
|
COMM domain containing 6 |
| chr2_-_233352531 | 0.92 |
ENST00000304546.1
|
ECEL1
|
endothelin converting enzyme-like 1 |
| chr4_-_80993717 | 0.92 |
ENST00000307333.7
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr16_+_30210552 | 0.91 |
ENST00000338971.5
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr8_+_77318769 | 0.90 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr2_-_101925055 | 0.90 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr3_+_127391769 | 0.90 |
ENST00000393363.3
ENST00000232744.8 ENST00000453791.2 |
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr22_-_29196511 | 0.89 |
ENST00000344347.5
|
XBP1
|
X-box binding protein 1 |
| chr11_+_129245796 | 0.89 |
ENST00000281437.4
|
BARX2
|
BARX homeobox 2 |
| chr1_-_154946825 | 0.88 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr19_+_6464502 | 0.87 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr4_-_177713788 | 0.87 |
ENST00000280193.2
|
VEGFC
|
vascular endothelial growth factor C |
| chr13_-_38443860 | 0.86 |
ENST00000426868.2
ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr3_-_13461807 | 0.86 |
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa |
| chr16_+_30212378 | 0.85 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr7_-_150675372 | 0.85 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr12_-_49318715 | 0.85 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr10_-_99531709 | 0.85 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr3_+_5020801 | 0.83 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr12_+_57482877 | 0.81 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr6_+_33359582 | 0.81 |
ENST00000450504.1
|
KIFC1
|
kinesin family member C1 |
| chr16_+_29472707 | 0.81 |
ENST00000565290.1
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr14_+_31343747 | 0.81 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr17_+_77751931 | 0.81 |
ENST00000310942.4
ENST00000269399.5 |
CBX2
|
chromobox homolog 2 |
| chr17_-_27278445 | 0.80 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr22_-_39096661 | 0.79 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chr22_-_39096925 | 0.79 |
ENST00000456626.1
ENST00000412832.1 |
JOSD1
|
Josephin domain containing 1 |
| chr1_+_110163709 | 0.78 |
ENST00000369840.2
ENST00000527846.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr22_-_39268192 | 0.78 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr14_+_31343951 | 0.78 |
ENST00000556908.1
ENST00000555881.1 ENST00000460581.2 |
COCH
|
cochlin |
| chr3_-_52312337 | 0.78 |
ENST00000469000.1
|
WDR82
|
WD repeat domain 82 |
| chr5_+_176784837 | 0.78 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr6_+_30524663 | 0.78 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr1_+_154947126 | 0.77 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr1_-_154946792 | 0.77 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr1_+_154947148 | 0.77 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr16_-_88752889 | 0.76 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr1_+_150522222 | 0.75 |
ENST00000369039.5
|
ADAMTSL4
|
ADAMTS-like 4 |
| chr5_+_141016969 | 0.75 |
ENST00000518856.1
|
RELL2
|
RELT-like 2 |
| chr1_-_201476220 | 0.74 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr22_-_29196546 | 0.74 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
| chr10_+_72432559 | 0.74 |
ENST00000373208.1
ENST00000373207.1 |
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr22_-_39096981 | 0.74 |
ENST00000427389.1
|
JOSD1
|
Josephin domain containing 1 |
| chr1_+_110163202 | 0.74 |
ENST00000531203.1
ENST00000256578.3 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr12_+_112451222 | 0.74 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr6_-_2245892 | 0.74 |
ENST00000380815.4
|
GMDS
|
GDP-mannose 4,6-dehydratase |
| chr4_-_11430221 | 0.72 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr17_+_40440481 | 0.71 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr19_+_41305406 | 0.71 |
ENST00000406058.2
ENST00000593726.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr1_+_150521876 | 0.70 |
ENST00000369041.5
ENST00000271643.4 ENST00000538795.1 |
ADAMTSL4
AL356356.1
|
ADAMTS-like 4 Protein LOC100996516 |
| chr17_-_6947225 | 0.70 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr1_+_110163682 | 0.69 |
ENST00000358729.4
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr19_-_42721819 | 0.69 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr15_-_41166414 | 0.69 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr19_-_10697895 | 0.68 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr7_-_132261253 | 0.67 |
ENST00000321063.4
|
PLXNA4
|
plexin A4 |
| chr14_+_56585048 | 0.67 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr1_+_110162448 | 0.67 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr17_-_62097927 | 0.67 |
ENST00000578313.1
ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2
|
intercellular adhesion molecule 2 |
| chrX_-_100914781 | 0.67 |
ENST00000431597.1
ENST00000458024.1 ENST00000413506.1 ENST00000440675.1 ENST00000328766.5 ENST00000356824.4 |
ARMCX2
|
armadillo repeat containing, X-linked 2 |
| chr19_+_1407733 | 0.66 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr19_+_41305612 | 0.65 |
ENST00000594380.1
ENST00000593397.1 ENST00000601733.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr3_-_128840604 | 0.65 |
ENST00000476465.1
ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43
|
RAB43, member RAS oncogene family |
| chr14_+_23564700 | 0.65 |
ENST00000554203.1
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chr17_+_40440094 | 0.65 |
ENST00000546010.2
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr3_-_194119083 | 0.65 |
ENST00000401815.1
|
GP5
|
glycoprotein V (platelet) |
| chr20_+_44098385 | 0.65 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr14_+_52164675 | 0.65 |
ENST00000555936.1
|
FRMD6
|
FERM domain containing 6 |
| chr22_-_32022280 | 0.64 |
ENST00000442379.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr9_-_136024721 | 0.64 |
ENST00000393160.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr1_+_29241027 | 0.64 |
ENST00000373797.1
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr19_-_10687907 | 0.63 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_+_41305627 | 0.62 |
ENST00000593525.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr7_-_132261223 | 0.61 |
ENST00000423507.2
|
PLXNA4
|
plexin A4 |
| chr14_+_56584414 | 0.61 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr20_+_44098346 | 0.61 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr3_-_107941230 | 0.60 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr1_-_54405773 | 0.60 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr1_+_226411319 | 0.60 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr14_+_90422239 | 0.59 |
ENST00000393452.3
ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr9_+_100000717 | 0.59 |
ENST00000375205.2
ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180
|
coiled-coil domain containing 180 |
| chr5_-_180230830 | 0.58 |
ENST00000427865.2
ENST00000514283.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr12_-_49365501 | 0.58 |
ENST00000403957.1
ENST00000301061.4 |
WNT10B
|
wingless-type MMTV integration site family, member 10B |
| chr15_-_79103757 | 0.57 |
ENST00000388820.4
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr8_-_74005349 | 0.57 |
ENST00000297354.6
|
SBSPON
|
somatomedin B and thrombospondin, type 1 domain containing |
| chrX_-_20284958 | 0.57 |
ENST00000379565.3
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr18_-_45935663 | 0.56 |
ENST00000589194.1
ENST00000591279.1 ENST00000590855.1 ENST00000587107.1 ENST00000588970.1 ENST00000586525.1 ENST00000592387.1 ENST00000590800.1 |
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr4_+_2420659 | 0.55 |
ENST00000382849.2
|
RP11-503N18.1
|
RP11-503N18.1 |
| chr19_+_41305740 | 0.55 |
ENST00000596517.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr16_+_67226019 | 0.55 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding |
| chr22_-_29196030 | 0.55 |
ENST00000405219.3
|
XBP1
|
X-box binding protein 1 |
| chr11_-_65308082 | 0.55 |
ENST00000532661.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr1_-_153949751 | 0.54 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr2_-_192015697 | 0.54 |
ENST00000409995.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr5_+_148206156 | 0.54 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.9 | 6.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.8 | 2.4 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.7 | 2.2 | GO:1903489 | epithelial cell differentiation involved in salivary gland development(GO:0060690) epithelial cell maturation involved in salivary gland development(GO:0060691) regulation of plasma cell differentiation(GO:1900098) positive regulation of plasma cell differentiation(GO:1900100) positive regulation of lactation(GO:1903489) |
| 0.7 | 4.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.6 | 1.9 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.6 | 4.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.6 | 0.6 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.6 | 1.7 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.5 | 2.5 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.5 | 1.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 5.0 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.4 | 3.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 1.3 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.4 | 1.2 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.4 | 1.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.3 | 2.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 | 3.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.3 | 2.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.3 | 1.3 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.3 | 1.3 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.3 | 2.5 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.3 | 2.4 | GO:0030578 | PML body organization(GO:0030578) |
| 0.3 | 2.6 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.3 | 1.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.3 | 1.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.3 | 0.9 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 2.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 1.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 3.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 1.8 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.2 | 0.7 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 4.0 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.2 | 1.4 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 1.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.9 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 0.7 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.2 | 1.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 0.5 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 0.5 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.2 | 0.5 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 1.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 2.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.6 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 1.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 1.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 3.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.6 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 1.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.8 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.7 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 2.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 1.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 3.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.5 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.7 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 1.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.5 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 5.6 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.1 | 0.4 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 1.2 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.4 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 | 0.3 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 1.7 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 1.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 1.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.8 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.5 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.7 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 1.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 1.7 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 1.7 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 1.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 2.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 1.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 1.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.5 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.6 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.6 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.0 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.2 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.9 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 3.5 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.8 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.8 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.7 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.5 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.8 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.4 | 3.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 6.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.4 | 1.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 4.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 4.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 1.7 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 1.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.2 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.2 | 0.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 7.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 1.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 1.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 4.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 3.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 4.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 1.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 3.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 9.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 1.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 2.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 1.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.4 | 1.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 6.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 1.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.3 | 1.8 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.3 | 2.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 1.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 1.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 1.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 1.3 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 3.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 1.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 1.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 1.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.5 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 2.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.6 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 1.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.6 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 6.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 7.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.6 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 1.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 2.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 2.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 1.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.5 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 3.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.1 | 1.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 2.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 4.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 3.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.4 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 2.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 5.2 | GO:0000988 | transcription factor activity, protein binding(GO:0000988) transcription factor activity, transcription factor binding(GO:0000989) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 2.6 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 8.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 6.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 7.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 3.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 4.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 4.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 8.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 3.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 5.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 3.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 2.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 4.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.1 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 2.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |