Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for HMX2
Z-value: 0.14
Transcription factors associated with HMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX2
|
ENSG00000188816.3 | H6 family homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX2 | hg19_v2_chr10_+_124907638_124907660 | 0.10 | 5.9e-01 | Click! |
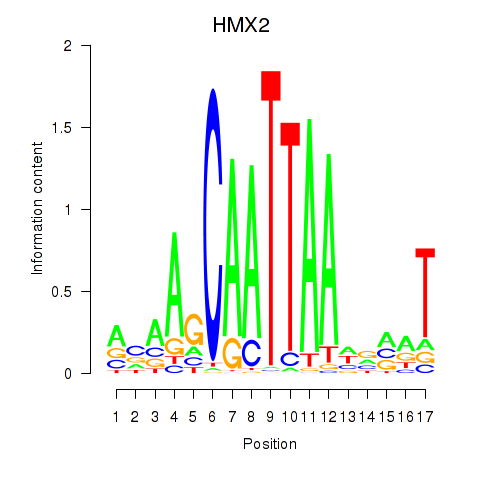
Activity profile of HMX2 motif
Sorted Z-values of HMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_150404904 | 3.64 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr1_+_201979645 | 3.60 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr4_-_155511887 | 3.28 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr2_-_89161432 | 3.11 |
ENST00000390242.2
|
IGKJ1
|
immunoglobulin kappa joining 1 |
| chr2_-_75745823 | 2.22 |
ENST00000452003.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr3_+_154801678 | 2.19 |
ENST00000462837.1
|
MME
|
membrane metallo-endopeptidase |
| chrX_+_105936982 | 2.08 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr17_-_46671323 | 2.01 |
ENST00000239151.5
|
HOXB5
|
homeobox B5 |
| chrX_+_105937068 | 2.00 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr4_-_100212132 | 1.81 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr15_+_58702742 | 1.78 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr2_+_211342432 | 1.74 |
ENST00000430249.2
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr15_+_41056218 | 1.73 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr6_+_25754927 | 1.72 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr17_-_64225508 | 1.68 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr1_+_56880606 | 1.67 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr3_-_194188956 | 1.61 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr4_-_69817481 | 1.52 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr1_+_209878182 | 1.48 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr6_-_135271219 | 1.39 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr15_+_41056255 | 1.35 |
ENST00000561160.1
ENST00000559445.1 |
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr2_+_234826016 | 1.32 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr15_+_69857515 | 1.29 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr2_-_151344172 | 1.29 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr3_-_187009646 | 1.28 |
ENST00000296280.6
ENST00000392470.2 ENST00000169293.6 ENST00000439271.1 ENST00000392472.2 ENST00000392475.2 |
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr2_-_160654745 | 1.28 |
ENST00000259053.4
ENST00000429078.2 |
CD302
|
CD302 molecule |
| chr16_+_82068585 | 1.27 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr6_+_25652432 | 1.26 |
ENST00000377961.2
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr3_-_187009798 | 1.25 |
ENST00000337774.5
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr2_+_211342400 | 1.21 |
ENST00000417946.1
ENST00000518043.1 ENST00000523702.1 |
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr1_+_196946680 | 1.20 |
ENST00000256785.4
|
CFHR5
|
complement factor H-related 5 |
| chr5_+_156607829 | 1.19 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr20_+_5986727 | 1.18 |
ENST00000378863.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_+_196946664 | 1.14 |
ENST00000367414.5
|
CFHR5
|
complement factor H-related 5 |
| chr3_+_122785895 | 1.14 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr13_+_49551020 | 1.14 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr10_-_12238071 | 1.12 |
ENST00000491614.1
ENST00000537776.1 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr11_-_327537 | 1.09 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr10_-_95241951 | 1.07 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr6_+_25652501 | 1.07 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr9_+_27109392 | 1.05 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr8_-_93978333 | 1.05 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_-_93978216 | 1.04 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_-_93978357 | 1.03 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr3_+_145782358 | 1.03 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr17_+_53016208 | 1.01 |
ENST00000574318.1
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr8_-_93978309 | 1.00 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr1_-_108231101 | 0.98 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr18_+_47087055 | 0.98 |
ENST00000577628.1
|
LIPG
|
lipase, endothelial |
| chr4_+_156775910 | 0.97 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr5_+_3596168 | 0.97 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr16_+_20685815 | 0.97 |
ENST00000561584.1
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr8_-_93978346 | 0.97 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr12_-_10151773 | 0.95 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr18_+_68002675 | 0.93 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr18_-_24765248 | 0.92 |
ENST00000580774.1
ENST00000284224.8 |
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr4_+_86748898 | 0.89 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_-_185726906 | 0.89 |
ENST00000513317.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr4_+_108745711 | 0.89 |
ENST00000394684.4
|
SGMS2
|
sphingomyelin synthase 2 |
| chr7_-_155604967 | 0.89 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr10_-_101945771 | 0.87 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr7_-_20256965 | 0.86 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr15_-_55700522 | 0.83 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr6_+_26124373 | 0.83 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr12_+_81471816 | 0.82 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr7_-_77325545 | 0.80 |
ENST00000447009.1
ENST00000416650.1 ENST00000440088.1 ENST00000430801.1 ENST00000398043.2 |
RSBN1L-AS1
|
RSBN1L antisense RNA 1 |
| chr11_-_118972575 | 0.80 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr4_+_86749045 | 0.79 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_+_48189612 | 0.79 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr19_-_36001113 | 0.76 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr9_-_75653627 | 0.75 |
ENST00000446946.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr2_-_190044480 | 0.73 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr21_-_34185944 | 0.73 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr11_-_26588634 | 0.73 |
ENST00000436318.2
ENST00000281268.8 |
MUC15
|
mucin 15, cell surface associated |
| chr4_+_166248775 | 0.72 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr17_-_70989072 | 0.71 |
ENST00000582769.1
|
SLC39A11
|
solute carrier family 39, member 11 |
| chr1_-_151804314 | 0.71 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr1_-_205290865 | 0.70 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr10_-_47239738 | 0.68 |
ENST00000413193.2
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr12_+_104680793 | 0.66 |
ENST00000529546.1
ENST00000529751.1 ENST00000540716.1 ENST00000528079.2 ENST00000526580.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr6_-_53481847 | 0.66 |
ENST00000503985.1
|
RP1-27K12.2
|
RP1-27K12.2 |
| chr12_+_104680659 | 0.66 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr12_-_94673956 | 0.64 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr13_+_24144509 | 0.64 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_45168875 | 0.63 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr10_-_95242044 | 0.63 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr3_-_187009468 | 0.63 |
ENST00000425937.1
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chrX_-_1331527 | 0.63 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chr8_+_31496809 | 0.61 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr11_+_112041253 | 0.60 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr2_+_158114051 | 0.58 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr12_-_10541575 | 0.58 |
ENST00000540818.1
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr20_+_5986756 | 0.57 |
ENST00000452938.1
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_+_144151520 | 0.57 |
ENST00000369372.4
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr4_-_39979576 | 0.55 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr16_-_20710212 | 0.54 |
ENST00000523065.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr1_-_217804377 | 0.52 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr15_-_50558223 | 0.52 |
ENST00000267845.3
|
HDC
|
histidine decarboxylase |
| chr15_-_55700457 | 0.52 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr3_+_133502877 | 0.51 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr10_-_22292613 | 0.50 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr14_-_106494587 | 0.49 |
ENST00000390597.2
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr4_-_26492076 | 0.49 |
ENST00000295589.3
|
CCKAR
|
cholecystokinin A receptor |
| chr18_+_72166564 | 0.48 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr17_-_295730 | 0.47 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr3_+_186560462 | 0.47 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr1_+_62439037 | 0.46 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr9_+_27109133 | 0.45 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr3_+_186560476 | 0.45 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr11_+_115498761 | 0.44 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr21_+_44073860 | 0.44 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chr3_-_121379739 | 0.44 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr2_+_118572226 | 0.44 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr7_-_7680601 | 0.43 |
ENST00000396682.2
|
RPA3
|
replication protein A3, 14kDa |
| chr17_-_72772462 | 0.42 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr20_+_3776371 | 0.42 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr11_+_60681346 | 0.41 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr11_-_18211042 | 0.41 |
ENST00000527671.1
|
RP11-113D6.6
|
Uncharacterized protein |
| chr19_+_9361606 | 0.41 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr3_-_46249878 | 0.41 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr6_-_51952418 | 0.40 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr14_+_64565442 | 0.40 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr10_-_126849626 | 0.40 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr1_+_99127225 | 0.38 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr22_-_39268308 | 0.38 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr2_-_55237484 | 0.37 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr11_+_128563652 | 0.37 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr16_-_21663919 | 0.37 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr19_+_999601 | 0.37 |
ENST00000594393.1
|
AC004528.1
|
Uncharacterized protein |
| chr9_+_131843377 | 0.36 |
ENST00000372546.4
ENST00000406974.3 ENST00000540102.1 |
DOLPP1
|
dolichyldiphosphatase 1 |
| chr11_+_110225855 | 0.36 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr16_-_21663950 | 0.36 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr19_-_3772209 | 0.35 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr10_+_99205894 | 0.35 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr1_+_186798073 | 0.34 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr16_+_19421803 | 0.34 |
ENST00000541464.1
|
TMC5
|
transmembrane channel-like 5 |
| chrX_+_119737806 | 0.34 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr19_-_41985048 | 0.34 |
ENST00000599801.1
ENST00000595063.1 ENST00000598215.1 |
AC011526.1
|
AC011526.1 |
| chr2_-_171571077 | 0.34 |
ENST00000409786.1
|
AC007405.2
|
long intergenic non-protein coding RNA 1124 |
| chr2_-_134326009 | 0.34 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr14_-_59950724 | 0.33 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr21_+_17909594 | 0.33 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr13_-_46756351 | 0.33 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr11_+_327171 | 0.32 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr21_+_44073916 | 0.32 |
ENST00000349112.3
ENST00000398224.3 |
PDE9A
|
phosphodiesterase 9A |
| chr10_+_99205959 | 0.32 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr4_+_90823130 | 0.31 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr6_-_151773232 | 0.31 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr11_-_64684672 | 0.30 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr2_-_20251744 | 0.30 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr10_-_115613828 | 0.29 |
ENST00000361384.2
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr8_+_32579341 | 0.29 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr14_-_50864006 | 0.29 |
ENST00000216378.2
|
CDKL1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr4_-_70518941 | 0.28 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr6_-_26199499 | 0.28 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr4_-_159080806 | 0.28 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr15_-_60695071 | 0.27 |
ENST00000557904.1
|
ANXA2
|
annexin A2 |
| chr19_-_3557570 | 0.27 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr1_-_110155671 | 0.27 |
ENST00000351050.3
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr15_+_35838446 | 0.27 |
ENST00000559210.1
|
DPH6-AS1
|
DPH6 antisense RNA 1 (head to head) |
| chr1_+_24019099 | 0.26 |
ENST00000443624.1
ENST00000458455.1 |
RPL11
|
ribosomal protein L11 |
| chrX_+_2670153 | 0.25 |
ENST00000509484.2
|
XG
|
Xg blood group |
| chr12_+_128399965 | 0.25 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr9_+_5890802 | 0.25 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr6_-_26199471 | 0.25 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr2_-_99279928 | 0.25 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr14_+_23564700 | 0.24 |
ENST00000554203.1
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chr2_+_172309634 | 0.24 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr6_-_2751146 | 0.24 |
ENST00000268446.5
ENST00000274643.7 |
MYLK4
|
myosin light chain kinase family, member 4 |
| chr17_-_72772425 | 0.24 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr8_-_71157595 | 0.23 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr12_+_107712173 | 0.23 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr5_+_167913450 | 0.22 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr19_-_14889349 | 0.22 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr17_-_73663245 | 0.21 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr1_-_115259337 | 0.21 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr6_+_114178512 | 0.21 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr2_-_166702601 | 0.21 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr6_-_26124138 | 0.21 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr7_-_84569561 | 0.20 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr3_+_179280668 | 0.20 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr1_+_192544857 | 0.20 |
ENST00000367459.3
ENST00000469578.2 |
RGS1
|
regulator of G-protein signaling 1 |
| chr10_-_126849588 | 0.20 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr2_+_1417228 | 0.20 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr11_+_94277017 | 0.20 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr17_-_47865948 | 0.20 |
ENST00000513602.1
|
FAM117A
|
family with sequence similarity 117, member A |
| chr10_-_75168071 | 0.19 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr1_+_70820451 | 0.19 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr17_-_38821373 | 0.19 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr2_+_27255806 | 0.19 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr5_-_134735568 | 0.19 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr5_-_114961673 | 0.19 |
ENST00000333314.3
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr1_-_63782888 | 0.18 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr19_+_12175504 | 0.18 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr5_-_43397184 | 0.18 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chrX_-_101186981 | 0.17 |
ENST00000458570.1
|
ZMAT1
|
zinc finger, matrin-type 1 |
| chr12_+_128399917 | 0.17 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr3_-_194119995 | 0.17 |
ENST00000323007.3
|
GP5
|
glycoprotein V (platelet) |
| chr10_-_22292675 | 0.16 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr2_+_201981663 | 0.16 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 1.0 | 3.0 | GO:1903718 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.5 | 3.6 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.3 | 0.9 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.3 | 0.9 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.3 | 4.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 1.0 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.2 | 1.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 0.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.0 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.2 | 3.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 0.6 | GO:0009946 | proximal/distal axis specification(GO:0009946) neuroblast differentiation(GO:0014016) |
| 0.2 | 3.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 2.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 3.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.8 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.2 | 0.6 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.2 | 1.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.2 | 1.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 0.5 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.2 | 2.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.5 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.5 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 1.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.8 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 1.7 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 1.8 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.2 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.9 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.8 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 1.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.9 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.9 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 1.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.3 | GO:0052360 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.2 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 1.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 1.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:2001189 | negative regulation of immunological synapse formation(GO:2000521) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.0 | 0.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 1.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.3 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 1.0 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 1.0 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 2.0 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 1.0 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.5 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 3.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.3 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 3.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 5.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.7 | 3.0 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.6 | 1.8 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.5 | 1.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.3 | 1.3 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.2 | 1.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 1.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 1.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 0.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 3.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.5 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.2 | 1.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.9 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.3 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.5 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 1.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 2.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.6 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 1.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 1.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 2.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 3.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 1.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 4.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 3.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |