Project
Illumina Body Map 2, young vs old
Navigation
Downloads
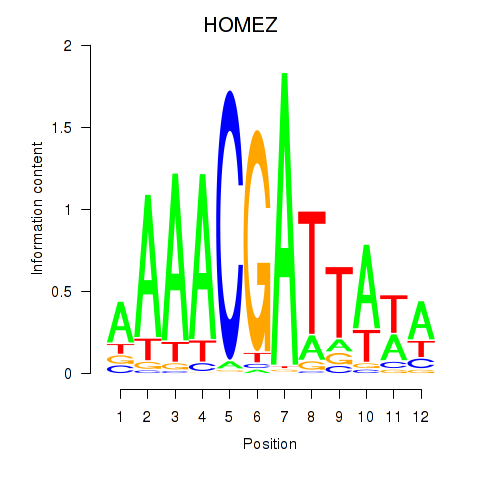
Results for HOMEZ
Z-value: 0.11
Transcription factors associated with HOMEZ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOMEZ
|
ENSG00000215271.6 | homeobox and leucine zipper encoding |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOMEZ | hg19_v2_chr14_-_23762777_23762821 | 0.53 | 1.6e-03 | Click! |
Activity profile of HOMEZ motif
Sorted Z-values of HOMEZ motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_202483867 | 3.19 |
ENST00000439802.1
ENST00000286195.3 ENST00000439140.1 ENST00000450242.1 |
ALS2CR11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 |
| chr19_-_22379753 | 2.60 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr12_-_10978957 | 2.33 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr6_+_30856507 | 2.27 |
ENST00000513240.1
ENST00000424544.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr9_-_95166976 | 2.16 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr2_+_79252822 | 2.12 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr11_-_63258601 | 2.08 |
ENST00000540857.1
ENST00000539221.1 ENST00000301790.4 |
HRASLS5
|
HRAS-like suppressor family, member 5 |
| chr19_+_44764031 | 1.93 |
ENST00000592581.1
ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233
|
zinc finger protein 233 |
| chr6_+_31105426 | 1.89 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chrX_-_92928557 | 1.85 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr16_+_33204156 | 1.80 |
ENST00000398667.4
|
TP53TG3C
|
TP53 target 3C |
| chr19_-_55677920 | 1.76 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr1_-_68516393 | 1.73 |
ENST00000395201.1
|
DIRAS3
|
DIRAS family, GTP-binding RAS-like 3 |
| chr9_-_95166884 | 1.68 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr14_+_101361107 | 1.68 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr2_-_61389168 | 1.64 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr8_-_52721975 | 1.64 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr3_-_196242233 | 1.63 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr9_+_71944241 | 1.63 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr21_+_17442799 | 1.60 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr7_+_99195677 | 1.60 |
ENST00000431679.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr7_+_116595028 | 1.58 |
ENST00000397751.1
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chrX_+_105192423 | 1.58 |
ENST00000540278.1
|
NRK
|
Nik related kinase |
| chr3_+_108308845 | 1.56 |
ENST00000479138.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr19_-_55677999 | 1.53 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr9_+_104296243 | 1.52 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr3_+_192958914 | 1.52 |
ENST00000264735.2
ENST00000602513.1 |
HRASLS
|
HRAS-like suppressor |
| chr2_+_11674213 | 1.50 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr15_+_63050785 | 1.48 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr2_+_79252804 | 1.48 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr12_-_64784471 | 1.45 |
ENST00000333722.5
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr4_-_101111615 | 1.41 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr19_-_55652290 | 1.38 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr5_+_175490540 | 1.37 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr2_+_79252834 | 1.33 |
ENST00000409471.1
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr5_+_140552218 | 1.32 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr1_+_202385953 | 1.32 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr19_-_46974741 | 1.32 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr6_-_74019870 | 1.31 |
ENST00000370384.3
|
KHDC1
|
KH homology domain containing 1 |
| chr9_-_95166841 | 1.30 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr9_+_104296163 | 1.30 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr1_-_205391178 | 1.30 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr12_+_75728419 | 1.26 |
ENST00000378695.4
ENST00000312442.2 |
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr15_+_23810903 | 1.24 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr19_-_18995029 | 1.23 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr10_+_53806501 | 1.22 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr22_-_22901636 | 1.21 |
ENST00000406503.1
ENST00000439106.1 ENST00000402697.1 ENST00000543184.1 ENST00000398743.2 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr18_+_44526744 | 1.20 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr12_+_26126681 | 1.20 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chrX_-_80457385 | 1.19 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr5_+_137203465 | 1.18 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr4_+_159131596 | 1.18 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr11_-_83878041 | 1.17 |
ENST00000398299.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr4_+_128702969 | 1.17 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr14_-_34420259 | 1.17 |
ENST00000250457.3
ENST00000547327.2 |
EGLN3
|
egl-9 family hypoxia-inducible factor 3 |
| chr9_+_124926856 | 1.17 |
ENST00000418632.1
|
MORN5
|
MORN repeat containing 5 |
| chr3_+_178276488 | 1.16 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr7_+_30589829 | 1.15 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr19_-_46974664 | 1.15 |
ENST00000438932.2
|
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr1_-_198509804 | 1.12 |
ENST00000489986.1
ENST00000367382.1 |
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
| chr1_+_119957554 | 1.11 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr2_-_238305397 | 1.11 |
ENST00000409809.1
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr6_-_137365402 | 1.11 |
ENST00000541547.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chr3_-_6847096 | 1.11 |
ENST00000454410.2
ENST00000424366.1 ENST00000417482.1 ENST00000412629.1 |
GRM7-AS3
|
GRM7 antisense RNA 3 |
| chr8_-_124749609 | 1.08 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr12_-_91573249 | 1.07 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr9_+_104296122 | 1.06 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr7_+_29874341 | 1.06 |
ENST00000409290.1
ENST00000242140.5 |
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr4_-_159956333 | 1.05 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr9_+_103235365 | 1.05 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr20_-_44168046 | 1.02 |
ENST00000372665.3
ENST00000372670.3 ENST00000600168.1 |
WFDC6
|
WAP four-disulfide core domain 6 |
| chr22_-_22901477 | 1.02 |
ENST00000420709.1
ENST00000398741.1 ENST00000405655.3 |
PRAME
|
preferentially expressed antigen in melanoma |
| chr7_+_6713376 | 1.02 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr4_+_169842707 | 1.01 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr9_-_138391692 | 1.01 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr5_-_110062384 | 0.99 |
ENST00000429839.2
|
TMEM232
|
transmembrane protein 232 |
| chr3_+_99536663 | 0.98 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chrX_-_114468605 | 0.98 |
ENST00000538422.1
ENST00000317135.8 |
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr5_+_7396141 | 0.98 |
ENST00000338316.4
|
ADCY2
|
adenylate cyclase 2 (brain) |
| chr6_-_37225367 | 0.97 |
ENST00000336655.2
|
TMEM217
|
transmembrane protein 217 |
| chr9_-_140142222 | 0.97 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr18_+_44526786 | 0.96 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr12_+_10365404 | 0.95 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr7_+_56032652 | 0.95 |
ENST00000437587.1
|
GBAS
|
glioblastoma amplified sequence |
| chr6_-_37225391 | 0.95 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr12_+_12223867 | 0.95 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr19_-_6433765 | 0.94 |
ENST00000321510.6
|
SLC25A41
|
solute carrier family 25, member 41 |
| chrX_+_49178536 | 0.93 |
ENST00000442437.2
|
GAGE12J
|
G antigen 12J |
| chr18_+_39739223 | 0.93 |
ENST00000601948.1
|
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr6_-_49937338 | 0.93 |
ENST00000398718.1
|
DEFB113
|
defensin, beta 113 |
| chr11_-_32816156 | 0.93 |
ENST00000531481.1
ENST00000335185.5 |
CCDC73
|
coiled-coil domain containing 73 |
| chr17_+_37856214 | 0.92 |
ENST00000445658.2
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr4_-_87278857 | 0.92 |
ENST00000509464.1
ENST00000511167.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr18_+_54814288 | 0.91 |
ENST00000585477.1
|
BOD1L2
|
biorientation of chromosomes in cell division 1-like 2 |
| chr12_-_112450915 | 0.90 |
ENST00000437003.2
ENST00000552374.2 ENST00000550831.3 ENST00000354825.3 ENST00000549537.2 ENST00000355445.3 |
TMEM116
|
transmembrane protein 116 |
| chr21_+_40824003 | 0.90 |
ENST00000452550.1
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr8_-_72274355 | 0.89 |
ENST00000388741.2
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr1_-_6420737 | 0.89 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr17_+_37856253 | 0.88 |
ENST00000540147.1
ENST00000584450.1 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr10_+_19777984 | 0.88 |
ENST00000377265.3
ENST00000455457.2 |
C10orf112
|
MAM and LDL receptor class A domain containing 1 |
| chr7_-_36406750 | 0.88 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr1_-_179545059 | 0.87 |
ENST00000367615.4
ENST00000367616.4 |
NPHS2
|
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr2_-_234475380 | 0.87 |
ENST00000443711.2
ENST00000251722.6 |
USP40
|
ubiquitin specific peptidase 40 |
| chr9_+_132962843 | 0.87 |
ENST00000458469.1
|
NCS1
|
neuronal calcium sensor 1 |
| chr1_+_163039143 | 0.86 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr2_+_1418154 | 0.86 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr4_+_95972822 | 0.86 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chrY_-_20935572 | 0.86 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr16_-_75467318 | 0.85 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr3_+_94657016 | 0.85 |
ENST00000462219.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr5_+_79331164 | 0.85 |
ENST00000350881.2
|
THBS4
|
thrombospondin 4 |
| chr12_+_19282643 | 0.83 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr4_+_3768075 | 0.82 |
ENST00000509482.1
ENST00000330055.5 |
ADRA2C
|
adrenoceptor alpha 2C |
| chr18_-_40857447 | 0.82 |
ENST00000590752.1
ENST00000596867.1 |
SYT4
|
synaptotagmin IV |
| chr3_-_179691866 | 0.81 |
ENST00000464614.1
ENST00000476138.1 ENST00000463761.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr1_-_169429690 | 0.81 |
ENST00000445428.1
|
RP1-206D15.3
|
RP1-206D15.3 |
| chr10_-_61122220 | 0.80 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr16_-_5115913 | 0.80 |
ENST00000474471.3
|
C16orf89
|
chromosome 16 open reading frame 89 |
| chr19_+_29704142 | 0.80 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr5_+_73109339 | 0.79 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr11_+_89867803 | 0.79 |
ENST00000321955.4
ENST00000525171.1 ENST00000375944.3 |
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr21_-_34863998 | 0.78 |
ENST00000402202.1
ENST00000381947.3 |
DNAJC28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr8_-_72274095 | 0.78 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr6_+_116601330 | 0.77 |
ENST00000449314.1
ENST00000453463.1 |
RP1-93H18.1
|
RP1-93H18.1 |
| chr8_-_98290087 | 0.77 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr8_+_39442097 | 0.77 |
ENST00000265707.5
ENST00000379866.1 ENST00000520772.1 ENST00000541111.1 |
ADAM18
|
ADAM metallopeptidase domain 18 |
| chr2_+_44502597 | 0.77 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chrY_+_20708557 | 0.77 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chrX_+_134124968 | 0.77 |
ENST00000330288.4
|
SMIM10
|
small integral membrane protein 10 |
| chr1_-_145076186 | 0.76 |
ENST00000369348.3
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr2_+_1488435 | 0.76 |
ENST00000446278.1
ENST00000469607.1 |
TPO
|
thyroid peroxidase |
| chr4_+_165878100 | 0.76 |
ENST00000513876.2
|
FAM218A
|
family with sequence similarity 218, member A |
| chr16_+_56226405 | 0.75 |
ENST00000565363.1
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr20_-_170317 | 0.75 |
ENST00000334391.4
|
DEFB128
|
defensin, beta 128 |
| chrX_+_49363665 | 0.74 |
ENST00000381700.6
|
GAGE1
|
G antigen 1 |
| chr7_-_18067478 | 0.74 |
ENST00000506618.2
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr4_-_87813566 | 0.73 |
ENST00000504008.1
ENST00000506308.1 |
C4orf36
|
chromosome 4 open reading frame 36 |
| chr18_+_11857439 | 0.73 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr4_+_5712942 | 0.72 |
ENST00000509451.1
|
EVC
|
Ellis van Creveld syndrome |
| chr3_+_108308559 | 0.72 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr1_-_60539422 | 0.72 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr2_+_29353520 | 0.72 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr16_-_5116025 | 0.72 |
ENST00000472572.3
ENST00000315997.5 ENST00000422873.1 ENST00000350219.4 |
C16orf89
|
chromosome 16 open reading frame 89 |
| chr5_+_140560980 | 0.72 |
ENST00000361016.2
|
PCDHB16
|
protocadherin beta 16 |
| chr6_+_125474992 | 0.71 |
ENST00000528193.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr8_-_112039643 | 0.71 |
ENST00000524283.1
|
RP11-946L20.2
|
RP11-946L20.2 |
| chr14_-_47812321 | 0.71 |
ENST00000357362.3
ENST00000486952.2 ENST00000426342.1 |
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr11_+_60467142 | 0.71 |
ENST00000529752.1
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr6_-_52628271 | 0.70 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr19_-_53636125 | 0.70 |
ENST00000601493.1
ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415
|
zinc finger protein 415 |
| chr22_+_19118321 | 0.70 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr5_+_1225470 | 0.70 |
ENST00000324642.3
ENST00000296821.4 |
SLC6A18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr21_-_34863693 | 0.70 |
ENST00000314399.3
|
DNAJC28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr1_+_202431859 | 0.69 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr6_+_29429217 | 0.69 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr10_+_24544249 | 0.69 |
ENST00000430453.2
|
KIAA1217
|
KIAA1217 |
| chr14_+_38264418 | 0.69 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr12_+_40019969 | 0.69 |
ENST00000405531.3
ENST00000398716.1 ENST00000324616.5 |
C12orf40
|
chromosome 12 open reading frame 40 |
| chr4_-_139051839 | 0.69 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr1_+_74701062 | 0.69 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chrX_-_33146477 | 0.69 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr4_+_96761238 | 0.68 |
ENST00000295266.4
|
PDHA2
|
pyruvate dehydrogenase (lipoamide) alpha 2 |
| chr8_+_7353368 | 0.68 |
ENST00000355602.2
|
DEFB107B
|
defensin, beta 107B |
| chr1_+_13910479 | 0.68 |
ENST00000509009.1
|
PDPN
|
podoplanin |
| chr6_+_53659746 | 0.68 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr3_-_100566492 | 0.67 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr21_+_17553910 | 0.67 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr21_+_22370717 | 0.67 |
ENST00000284894.7
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr18_-_31802056 | 0.66 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr4_+_83956237 | 0.66 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr17_-_42906965 | 0.66 |
ENST00000586267.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr4_+_169753156 | 0.66 |
ENST00000393726.3
ENST00000507735.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_+_210517895 | 0.66 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chrX_+_49235708 | 0.65 |
ENST00000381725.1
|
GAGE2B
|
G antigen 2B |
| chr1_-_100643765 | 0.65 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr14_-_92198403 | 0.65 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr17_+_37856299 | 0.64 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr9_+_135937365 | 0.64 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chrX_+_36254051 | 0.64 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr12_-_71551652 | 0.64 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chrX_+_49160148 | 0.64 |
ENST00000407599.3
|
GAGE10
|
G antigen 10 |
| chr5_+_95066823 | 0.63 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr3_+_44754126 | 0.63 |
ENST00000449836.1
ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr1_-_113161730 | 0.63 |
ENST00000544629.1
ENST00000543570.1 ENST00000360743.4 ENST00000490067.1 ENST00000343210.7 ENST00000369666.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr5_-_159827033 | 0.63 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr8_-_13372253 | 0.63 |
ENST00000316609.5
|
DLC1
|
deleted in liver cancer 1 |
| chr1_+_163038565 | 0.62 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr12_+_10365082 | 0.62 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr12_-_52911718 | 0.62 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr5_-_159827073 | 0.62 |
ENST00000408953.3
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chrX_+_86772707 | 0.62 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr4_+_144312659 | 0.62 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr19_-_53400813 | 0.62 |
ENST00000595635.1
ENST00000594741.1 ENST00000597111.1 ENST00000593618.1 ENST00000597909.1 |
ZNF320
|
zinc finger protein 320 |
| chr18_-_19994830 | 0.61 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr4_-_46126093 | 0.61 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr8_+_86019382 | 0.61 |
ENST00000360375.3
|
LRRCC1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr4_+_83956312 | 0.61 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr3_-_107941209 | 0.60 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr3_-_126327398 | 0.60 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr16_-_29910365 | 0.60 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOMEZ
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.5 | 1.5 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.4 | 1.3 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.4 | 1.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.4 | 1.6 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.4 | 3.9 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.3 | 0.8 | GO:0048174 | negative regulation of short-term neuronal synaptic plasticity(GO:0048174) |
| 0.2 | 2.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 1.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.7 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 0.9 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 0.8 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 4.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 0.4 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.2 | 1.0 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.2 | 0.5 | GO:0019254 | amino-acid betaine catabolic process(GO:0006579) carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.9 | GO:0072248 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.2 | 1.9 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 1.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.7 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 2.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.4 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.4 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.5 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.8 | GO:0015744 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) succinate transmembrane transport(GO:0071422) |
| 0.1 | 0.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.8 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.7 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 2.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.5 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 2.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 4.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.6 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.7 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.5 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.8 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 3.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 1.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.5 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.4 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.6 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 1.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.9 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.2 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 1.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.5 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 1.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 2.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.2 | GO:1902102 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 1.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.4 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.9 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 0.6 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.3 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 1.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 1.1 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 1.7 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 1.5 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 1.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.3 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.7 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.5 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.4 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 1.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.2 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 1.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 1.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.0 | 0.8 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.2 | GO:2000197 | regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 1.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.3 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 1.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.6 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.7 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 1.0 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.8 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 3.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.4 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.6 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.4 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.4 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.0 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 1.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 2.8 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 2.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.8 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.6 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.6 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.3 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 1.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.0 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 5.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.8 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 5.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 1.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.3 | 0.8 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.3 | 4.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 0.7 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 2.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 1.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 2.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 0.5 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 0.7 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 2.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.6 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.4 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 2.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.8 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.4 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.7 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.5 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.3 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 2.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.7 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.8 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.6 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 1.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.5 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 1.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 0.9 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.6 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.8 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 1.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 1.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 2.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.7 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.3 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 4.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 1.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 5.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 4.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.0 | GO:0015563 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.0 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.8 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 4.6 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.8 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 3.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 3.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |