Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for HOXA9
Z-value: 0.58
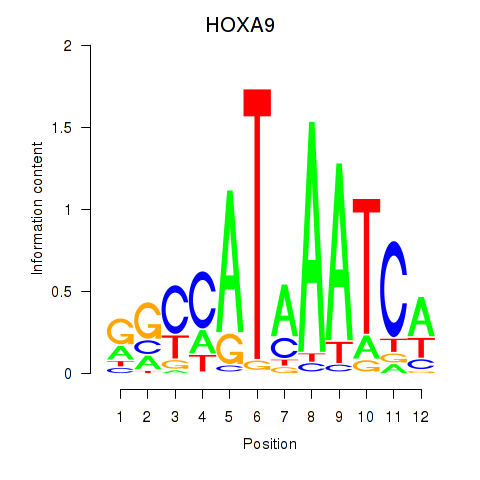
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.11 | homeobox A9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg19_v2_chr7_-_27205136_27205164 | 0.25 | 1.7e-01 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_88427568 | 4.59 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr2_-_224467002 | 3.49 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr13_+_53602894 | 2.74 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr1_+_50575292 | 2.70 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_-_224467093 | 2.63 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chrX_+_65384182 | 2.37 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr2_-_154335300 | 2.36 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr15_+_33022885 | 2.35 |
ENST00000322805.4
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr10_+_85933494 | 2.26 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr7_-_92855762 | 2.16 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr8_+_76452097 | 2.16 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr7_+_54609995 | 2.15 |
ENST00000302287.3
ENST00000407838.3 |
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr1_-_204135450 | 1.98 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chrX_-_13835398 | 1.87 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr19_-_47975417 | 1.74 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr3_+_120626919 | 1.64 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chr11_-_35441597 | 1.63 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_-_69536346 | 1.62 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr1_+_24645807 | 1.60 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24645865 | 1.57 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr11_-_35441524 | 1.57 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_-_71176623 | 1.56 |
ENST00000373306.4
|
TACR2
|
tachykinin receptor 2 |
| chr17_-_29624343 | 1.54 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chrX_-_13835147 | 1.49 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr12_-_71148413 | 1.48 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr11_-_35440796 | 1.46 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr1_+_50574585 | 1.45 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chrX_+_103031758 | 1.43 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr14_+_101361107 | 1.41 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr7_+_20370746 | 1.41 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr5_-_146833222 | 1.40 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr15_-_41166414 | 1.40 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chrX_-_13835461 | 1.39 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr4_-_69434245 | 1.37 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chrX_-_92928557 | 1.35 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr11_+_27062502 | 1.33 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr11_+_27062860 | 1.32 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr20_+_58203664 | 1.25 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chrX_+_65384052 | 1.24 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr14_+_78870030 | 1.23 |
ENST00000553631.1
ENST00000554719.1 |
NRXN3
|
neurexin 3 |
| chr5_-_146833485 | 1.22 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr10_+_15001430 | 1.21 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chrX_+_65382433 | 1.20 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr10_-_134599556 | 1.19 |
ENST00000368592.5
|
NKX6-2
|
NK6 homeobox 2 |
| chr11_-_126870655 | 1.19 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr11_-_126870683 | 1.16 |
ENST00000525704.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr2_-_209051727 | 1.16 |
ENST00000453017.1
ENST00000423952.2 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr6_+_160327974 | 1.15 |
ENST00000252660.4
|
MAS1
|
MAS1 oncogene |
| chr3_+_148545586 | 1.12 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr1_+_101185290 | 1.11 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr5_-_131347583 | 1.10 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr7_-_16505440 | 1.10 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr18_-_10701979 | 1.09 |
ENST00000538948.1
ENST00000285141.4 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr17_-_66951474 | 1.08 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr7_+_119913688 | 1.07 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr7_-_83824169 | 1.06 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr4_-_176733377 | 1.05 |
ENST00000505375.1
|
GPM6A
|
glycoprotein M6A |
| chr11_-_17555421 | 1.04 |
ENST00000526181.1
|
USH1C
|
Usher syndrome 1C (autosomal recessive, severe) |
| chr4_+_113970772 | 1.03 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr7_-_83824449 | 1.02 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_7493390 | 1.00 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr9_+_77112244 | 0.99 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr15_+_59730348 | 0.99 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr5_+_140602904 | 0.99 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr19_+_46367518 | 0.98 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr3_-_47622282 | 0.95 |
ENST00000383738.2
ENST00000264723.4 |
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C) |
| chr8_-_28347737 | 0.93 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr5_-_131347501 | 0.93 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr10_-_96829246 | 0.92 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr8_-_71157595 | 0.92 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr2_+_108994633 | 0.91 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr15_+_40643227 | 0.91 |
ENST00000448599.2
|
PHGR1
|
proline/histidine/glycine-rich 1 |
| chr10_+_18689637 | 0.90 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_+_192544857 | 0.89 |
ENST00000367459.3
ENST00000469578.2 |
RGS1
|
regulator of G-protein signaling 1 |
| chr10_+_51549498 | 0.89 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr3_+_63428752 | 0.89 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr8_-_20831355 | 0.89 |
ENST00000518967.1
|
RP11-369E15.3
|
RP11-369E15.3 |
| chr7_-_14880892 | 0.88 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr17_+_48912744 | 0.88 |
ENST00000311378.4
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr2_+_232063260 | 0.87 |
ENST00000349938.4
|
ARMC9
|
armadillo repeat containing 9 |
| chr16_+_15596123 | 0.83 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr19_-_18314836 | 0.83 |
ENST00000464076.3
ENST00000222256.4 |
RAB3A
|
RAB3A, member RAS oncogene family |
| chr11_+_27062272 | 0.83 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr11_+_1430983 | 0.82 |
ENST00000524702.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr10_+_60759378 | 0.82 |
ENST00000432535.1
|
LINC00844
|
long intergenic non-protein coding RNA 844 |
| chr10_+_81967456 | 0.82 |
ENST00000422847.1
|
LINC00857
|
long intergenic non-protein coding RNA 857 |
| chr4_-_110723134 | 0.82 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr14_+_38264418 | 0.82 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr3_+_26664291 | 0.81 |
ENST00000396641.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr6_+_24126350 | 0.80 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr7_+_153584166 | 0.80 |
ENST00000404039.1
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr8_+_16884740 | 0.79 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr1_+_149230680 | 0.79 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chr17_+_63133587 | 0.78 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr8_+_69242957 | 0.77 |
ENST00000518698.1
ENST00000539993.1 |
C8orf34
|
chromosome 8 open reading frame 34 |
| chr1_+_24646263 | 0.77 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr10_+_124320156 | 0.76 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr2_+_149974684 | 0.76 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr4_-_99578776 | 0.76 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr9_-_127269661 | 0.75 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr6_+_99282570 | 0.75 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr10_+_102222798 | 0.75 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr17_+_60447579 | 0.74 |
ENST00000450662.2
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr1_+_24646002 | 0.74 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr10_-_79397547 | 0.74 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_-_125351481 | 0.74 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr16_-_65106110 | 0.73 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr6_-_84417633 | 0.73 |
ENST00000521931.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr4_-_99578789 | 0.73 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chrX_-_72434628 | 0.72 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr6_-_88876058 | 0.71 |
ENST00000369501.2
|
CNR1
|
cannabinoid receptor 1 (brain) |
| chr4_+_154074217 | 0.70 |
ENST00000437508.2
|
TRIM2
|
tripartite motif containing 2 |
| chr17_+_54230819 | 0.70 |
ENST00000318698.2
ENST00000566473.2 |
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr12_+_15475462 | 0.69 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr2_+_149894968 | 0.68 |
ENST00000409642.3
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr1_+_166958346 | 0.68 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr6_+_25754927 | 0.68 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr14_+_88490894 | 0.68 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chrX_+_144908928 | 0.68 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr9_+_21440440 | 0.67 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr6_-_25726781 | 0.67 |
ENST00000297012.3
|
HIST1H2AA
|
histone cluster 1, H2aa |
| chr1_+_24645915 | 0.66 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr2_-_225811747 | 0.65 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr4_-_110723194 | 0.65 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr1_-_166028709 | 0.64 |
ENST00000595430.1
|
AL626787.1
|
AL626787.1 |
| chr12_-_57444957 | 0.64 |
ENST00000433964.1
|
MYO1A
|
myosin IA |
| chr8_+_35649365 | 0.64 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr13_-_47471155 | 0.64 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr6_+_126240442 | 0.64 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr16_-_67427389 | 0.63 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr10_-_90712520 | 0.63 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr5_+_140480083 | 0.63 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr1_-_38512450 | 0.63 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr3_-_143567262 | 0.63 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr9_-_95640218 | 0.62 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr5_-_43515231 | 0.62 |
ENST00000306862.2
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chrX_+_65382381 | 0.62 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr13_+_98794810 | 0.62 |
ENST00000595437.1
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr16_+_70613770 | 0.61 |
ENST00000429149.2
ENST00000563721.2 |
IL34
|
interleukin 34 |
| chr17_-_8021710 | 0.61 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr8_+_24151620 | 0.60 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr12_+_15475331 | 0.60 |
ENST00000281171.4
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr8_-_20040638 | 0.59 |
ENST00000519026.1
ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr17_+_63133526 | 0.59 |
ENST00000443584.3
|
RGS9
|
regulator of G-protein signaling 9 |
| chr9_-_104198042 | 0.59 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr3_+_190917023 | 0.59 |
ENST00000445281.1
|
OSTN
|
osteocrin |
| chr1_-_72748140 | 0.59 |
ENST00000434200.1
|
NEGR1
|
neuronal growth regulator 1 |
| chr3_-_58613323 | 0.58 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr7_-_99277610 | 0.58 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr12_-_74796291 | 0.57 |
ENST00000551726.1
|
RP11-81H3.2
|
RP11-81H3.2 |
| chr1_-_198509804 | 0.57 |
ENST00000489986.1
ENST00000367382.1 |
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
| chr5_+_126984710 | 0.56 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr6_+_47666275 | 0.56 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr22_+_32754139 | 0.56 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chrX_+_36246735 | 0.55 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr4_-_16085657 | 0.54 |
ENST00000543373.1
|
PROM1
|
prominin 1 |
| chr7_+_117251671 | 0.54 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr6_+_71104588 | 0.54 |
ENST00000418403.1
|
RP11-462G2.1
|
RP11-462G2.1 |
| chr4_-_176733897 | 0.54 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr3_+_149192475 | 0.53 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr6_-_55443958 | 0.52 |
ENST00000370850.2
|
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr2_+_138721850 | 0.52 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr4_-_186732048 | 0.52 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_21368075 | 0.52 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr10_-_128210005 | 0.52 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr2_+_108994466 | 0.50 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr5_+_38845960 | 0.50 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr20_-_17539456 | 0.50 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chrX_+_114827818 | 0.50 |
ENST00000420625.2
|
PLS3
|
plastin 3 |
| chr6_-_32374900 | 0.50 |
ENST00000374995.3
ENST00000374993.1 ENST00000414363.1 ENST00000540315.1 ENST00000544175.1 ENST00000429232.2 ENST00000454136.3 ENST00000446536.2 |
BTNL2
|
butyrophilin-like 2 (MHC class II associated) |
| chr3_-_72149553 | 0.49 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr10_+_5488564 | 0.48 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr12_-_91573249 | 0.48 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr1_+_150254936 | 0.47 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr12_+_93130311 | 0.47 |
ENST00000344636.3
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr8_+_24151553 | 0.47 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr17_-_56494908 | 0.47 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr5_+_38846101 | 0.47 |
ENST00000274276.3
|
OSMR
|
oncostatin M receptor |
| chr2_-_31637560 | 0.46 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr6_-_131299929 | 0.46 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr16_-_18462221 | 0.46 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr1_+_104615595 | 0.46 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr19_-_14945933 | 0.46 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr6_-_55443831 | 0.45 |
ENST00000428842.1
ENST00000358072.5 ENST00000508459.1 |
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr5_-_131347306 | 0.45 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr9_+_40028620 | 0.45 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr3_+_178276488 | 0.45 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr15_+_27216297 | 0.45 |
ENST00000333743.6
|
GABRG3
|
gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
| chr5_-_148758839 | 0.45 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr6_+_25727046 | 0.45 |
ENST00000274764.2
|
HIST1H2BA
|
histone cluster 1, H2ba |
| chr17_+_57297807 | 0.43 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr5_-_110062349 | 0.43 |
ENST00000511883.2
ENST00000455884.2 |
TMEM232
|
transmembrane protein 232 |
| chr17_-_31404 | 0.43 |
ENST00000343572.7
|
DOC2B
|
double C2-like domains, beta |
| chr10_-_28270795 | 0.42 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chrX_-_110513703 | 0.41 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr10_-_45474237 | 0.41 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr8_+_38261880 | 0.41 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr7_+_93535866 | 0.41 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr2_+_232063436 | 0.40 |
ENST00000440107.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr15_+_59664884 | 0.40 |
ENST00000558348.1
|
FAM81A
|
family with sequence similarity 81, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.8 | 2.4 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 | 2.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.4 | 4.8 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 2.2 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.4 | 1.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 4.7 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 1.2 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.4 | 6.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 1.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.3 | 2.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.3 | 1.6 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.3 | 0.9 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.3 | 0.8 | GO:1990709 | maintenance of synapse structure(GO:0099558) presynaptic active zone organization(GO:1990709) |
| 0.3 | 1.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 2.7 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.3 | 4.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 3.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.0 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.2 | 1.7 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 5.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 1.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 1.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 5.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 1.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 0.8 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.7 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.5 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.6 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 3.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 1.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.6 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.1 | 0.5 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.9 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.5 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 0.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.8 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) regulation of stem cell division(GO:2000035) |
| 0.1 | 0.4 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 1.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.6 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.6 | GO:0001578 | microtubule bundle formation(GO:0001578) microtubule polymerization(GO:0046785) |
| 0.1 | 0.3 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 1.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 2.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 3.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 1.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 1.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.7 | GO:0034465 | response to carbon monoxide(GO:0034465) smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 1.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.1 | 1.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.7 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 2.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.3 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.5 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 1.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.7 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 1.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.8 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.3 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.8 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 1.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.8 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.1 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 1.4 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.1 | GO:1900020 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 1.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.5 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.1 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.0 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.0 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.4 | 1.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.3 | 1.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.3 | 0.8 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.3 | 4.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 6.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.7 | GO:0030849 | autosome(GO:0030849) |
| 0.2 | 0.9 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.5 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 3.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 2.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 2.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 3.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.7 | 4.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 4.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 5.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.3 | 1.6 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 1.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 3.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 0.9 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 1.0 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.2 | 0.9 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 0.9 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.2 | 2.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 1.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.4 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.5 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 1.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.6 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 0.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 1.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 6.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.9 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 1.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.4 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 4.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 1.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 2.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 3.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 2.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.5 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.6 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 2.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.8 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 2.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 2.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 2.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 5.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 3.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 1.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 2.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 4.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 4.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |