Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for HOXB3
Z-value: 0.53
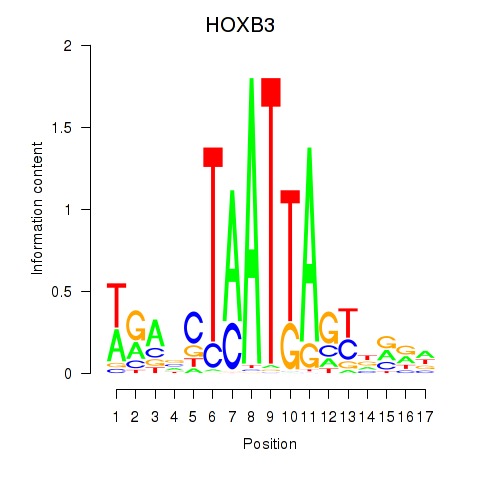
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.7 | homeobox B3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB3 | hg19_v2_chr17_-_46667594_46667619 | 0.32 | 7.8e-02 | Click! |
Activity profile of HOXB3 motif
Sorted Z-values of HOXB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_89160770 | 7.43 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr2_-_89161432 | 7.19 |
ENST00000390242.2
|
IGKJ1
|
immunoglobulin kappa joining 1 |
| chr2_-_89161064 | 6.47 |
ENST00000390241.2
|
IGKJ2
|
immunoglobulin kappa joining 2 |
| chr12_-_123201337 | 5.46 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr15_-_44969022 | 5.14 |
ENST00000560110.1
|
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr14_-_106494587 | 4.91 |
ENST00000390597.2
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr12_-_123187890 | 4.31 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr2_+_90043607 | 4.26 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr2_-_89476644 | 4.12 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr1_+_81106951 | 3.91 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr21_+_43823983 | 3.87 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr19_+_17638059 | 3.62 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr14_+_22564294 | 3.52 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr22_+_22730353 | 3.50 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr14_+_22465771 | 3.24 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr2_+_89999259 | 3.22 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr15_-_44969086 | 3.22 |
ENST00000434130.1
ENST00000560780.1 |
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr2_+_89184868 | 3.19 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr11_-_118083600 | 3.17 |
ENST00000524477.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr4_-_25865159 | 2.97 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chrX_+_135730373 | 2.94 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr2_+_90077680 | 2.92 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr14_-_107179265 | 2.76 |
ENST00000390634.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr19_+_17638041 | 2.74 |
ENST00000601861.1
|
FAM129C
|
family with sequence similarity 129, member C |
| chr4_+_40198527 | 2.71 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chrX_+_135730297 | 2.71 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr3_+_114012819 | 2.70 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr7_+_142353445 | 2.68 |
ENST00000390396.1
|
TRBV23-1
|
T cell receptor beta variable 23-1 (non-functional) |
| chr16_+_12059050 | 2.66 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr14_+_22180536 | 2.51 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr1_-_206671061 | 2.25 |
ENST00000367119.1
|
C1orf147
|
chromosome 1 open reading frame 147 |
| chr1_+_222910538 | 2.24 |
ENST00000434700.1
ENST00000445590.2 |
FAM177B
|
family with sequence similarity 177, member B |
| chr16_+_31366536 | 2.16 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr3_-_151047327 | 2.16 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr14_+_22320634 | 2.13 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr11_-_107729287 | 2.09 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_-_39321512 | 2.08 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chrX_-_70326455 | 2.04 |
ENST00000374251.5
|
CXorf65
|
chromosome X open reading frame 65 |
| chr14_-_106926724 | 2.03 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr11_-_107729504 | 2.01 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_+_87769459 | 2.00 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr22_-_32651326 | 1.90 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr1_+_222910625 | 1.86 |
ENST00000360827.2
|
FAM177B
|
family with sequence similarity 177, member B |
| chr11_-_107729887 | 1.85 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_-_8088773 | 1.85 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr11_+_63273811 | 1.83 |
ENST00000340246.5
|
LGALS12
|
lectin, galactoside-binding, soluble, 12 |
| chr3_+_121774202 | 1.82 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr4_+_86525299 | 1.81 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_+_72595971 | 1.77 |
ENST00000581412.1
|
CTD-2006K23.1
|
CTD-2006K23.1 |
| chr18_+_61554932 | 1.71 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr14_+_61654271 | 1.60 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr3_-_130745403 | 1.58 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr2_-_112237835 | 1.57 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr17_-_8770956 | 1.53 |
ENST00000311434.9
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6 |
| chr6_-_116866773 | 1.52 |
ENST00000368602.3
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr11_-_104827425 | 1.49 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr8_+_92261516 | 1.49 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr20_+_58571419 | 1.47 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr12_-_8088871 | 1.47 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr15_-_55562479 | 1.43 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_-_104905840 | 1.43 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr15_-_55563072 | 1.42 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_-_54824344 | 1.38 |
ENST00000346508.3
ENST00000446712.3 ENST00000432233.3 ENST00000301219.3 |
LILRA5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr1_+_161691353 | 1.37 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr7_-_92777606 | 1.37 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr17_+_62223320 | 1.36 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr16_+_50730910 | 1.36 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr6_-_62996066 | 1.35 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr15_-_55562582 | 1.35 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_56522001 | 1.29 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr12_+_56521840 | 1.27 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr14_+_22947861 | 1.27 |
ENST00000390482.1
|
TRAJ57
|
T cell receptor alpha joining 57 |
| chr11_-_104817919 | 1.26 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr18_+_61557781 | 1.25 |
ENST00000443281.1
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr9_+_125132803 | 1.22 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr15_-_55562451 | 1.21 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_+_5710919 | 1.20 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr17_-_8113542 | 1.20 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr17_-_62208169 | 1.18 |
ENST00000606895.1
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr5_+_148737562 | 1.15 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr12_-_10022735 | 1.14 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr1_-_21620877 | 1.13 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr11_-_26593677 | 1.13 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr19_+_1077393 | 1.11 |
ENST00000590577.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr21_-_15918618 | 1.11 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr20_-_56647116 | 1.11 |
ENST00000441277.2
ENST00000452842.1 |
RP13-379L11.1
|
RP13-379L11.1 |
| chr6_+_130339710 | 1.09 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr17_-_73258425 | 1.08 |
ENST00000578348.1
ENST00000582486.1 ENST00000582717.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr12_+_66582919 | 1.08 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr9_+_125133315 | 1.05 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_-_33102338 | 1.04 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr14_+_100485712 | 1.04 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr1_+_158978768 | 1.04 |
ENST00000447473.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr7_-_33102399 | 1.03 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr7_+_30589829 | 1.03 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr8_+_92114060 | 1.01 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr12_+_55248289 | 1.01 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr16_-_21431078 | 1.00 |
ENST00000458643.2
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr19_+_49990811 | 0.99 |
ENST00000391857.4
ENST00000467825.2 |
RPL13A
|
ribosomal protein L13a |
| chr11_-_13517565 | 0.99 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr4_-_111563279 | 0.97 |
ENST00000511837.1
|
PITX2
|
paired-like homeodomain 2 |
| chr2_-_101925055 | 0.97 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr2_+_143635222 | 0.95 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr8_+_27169138 | 0.93 |
ENST00000522338.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr10_-_4285835 | 0.92 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr9_-_125590818 | 0.92 |
ENST00000259467.4
|
PDCL
|
phosducin-like |
| chr11_+_47293795 | 0.91 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr4_-_40632881 | 0.90 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chrX_-_21676442 | 0.89 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr15_+_64680003 | 0.89 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr14_+_23002427 | 0.86 |
ENST00000390527.1
|
TRAJ10
|
T cell receptor alpha joining 10 |
| chr14_+_56584414 | 0.85 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr7_-_55620433 | 0.85 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr11_-_96076334 | 0.85 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr14_-_24711764 | 0.84 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr6_+_26501449 | 0.83 |
ENST00000244513.6
|
BTN1A1
|
butyrophilin, subfamily 1, member A1 |
| chr2_+_172309634 | 0.83 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr11_+_121447469 | 0.83 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr4_-_164534657 | 0.82 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr13_-_30160925 | 0.81 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr16_+_12059091 | 0.81 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr3_-_112564797 | 0.81 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr7_-_99716914 | 0.80 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr14_+_57671888 | 0.80 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr9_-_3489406 | 0.79 |
ENST00000457373.1
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr6_+_26045603 | 0.78 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr11_-_124981475 | 0.78 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr4_-_109541539 | 0.78 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr18_-_44181442 | 0.77 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr12_-_10605929 | 0.77 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr4_-_40632844 | 0.77 |
ENST00000505414.1
|
RBM47
|
RNA binding motif protein 47 |
| chr9_+_131062367 | 0.76 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr17_-_57229155 | 0.75 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr19_+_9296279 | 0.75 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr14_-_24711470 | 0.75 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr4_-_111563076 | 0.74 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr4_-_120243545 | 0.74 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr16_-_21868739 | 0.73 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr19_-_3985455 | 0.73 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr3_-_130745571 | 0.73 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr1_+_28261492 | 0.72 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr4_-_74486109 | 0.72 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr1_-_21625486 | 0.71 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr2_+_138722028 | 0.69 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr17_-_40337470 | 0.69 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr20_-_55100981 | 0.68 |
ENST00000243913.4
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr13_+_73632897 | 0.68 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr1_+_46972668 | 0.67 |
ENST00000371956.4
ENST00000360032.3 |
DMBX1
|
diencephalon/mesencephalon homeobox 1 |
| chr2_+_143635067 | 0.67 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr4_-_19458597 | 0.67 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr8_+_27168988 | 0.67 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr11_-_59950622 | 0.66 |
ENST00000323961.3
ENST00000412309.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr16_-_21868978 | 0.66 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr19_+_48949030 | 0.66 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr1_+_24019099 | 0.66 |
ENST00000443624.1
ENST00000458455.1 |
RPL11
|
ribosomal protein L11 |
| chrX_+_77166172 | 0.65 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr14_-_24711806 | 0.64 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_-_185538849 | 0.63 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr7_+_129015484 | 0.63 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr8_-_623547 | 0.62 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr1_+_192605252 | 0.62 |
ENST00000391995.2
ENST00000543215.1 |
RGS13
|
regulator of G-protein signaling 13 |
| chr5_+_150639360 | 0.62 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr16_-_30257122 | 0.61 |
ENST00000520915.1
|
RP11-347C12.1
|
Putative NPIP-like protein LOC613037 |
| chrX_+_10031499 | 0.61 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr1_+_79115503 | 0.61 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr2_-_70780572 | 0.61 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr16_-_21436459 | 0.61 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr14_-_24711865 | 0.61 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr7_+_99717230 | 0.60 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr17_-_44657017 | 0.59 |
ENST00000573185.1
ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A
|
ADP-ribosylation factor-like 17A |
| chr19_+_48949087 | 0.59 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr11_-_26593779 | 0.59 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr19_+_36632485 | 0.59 |
ENST00000586963.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr2_+_28618532 | 0.59 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr8_+_32579271 | 0.58 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr11_-_59950486 | 0.57 |
ENST00000426738.2
ENST00000533023.1 ENST00000420732.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr16_-_29415350 | 0.56 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr7_-_99764907 | 0.56 |
ENST00000413800.1
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4 |
| chr14_-_90085458 | 0.56 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr16_-_3350614 | 0.55 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr2_+_172778952 | 0.55 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr1_+_159750776 | 0.54 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr11_-_63439013 | 0.54 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr11_-_59950519 | 0.53 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr6_-_32095968 | 0.53 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr19_-_3557570 | 0.53 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr1_-_1709845 | 0.53 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr7_+_5919458 | 0.52 |
ENST00000416608.1
|
OCM
|
oncomodulin |
| chr5_+_140019004 | 0.52 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr17_-_10372875 | 0.52 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr17_-_44439084 | 0.52 |
ENST00000575960.1
ENST00000575698.1 ENST00000571246.1 ENST00000434041.2 ENST00000570618.1 ENST00000450673.3 |
ARL17B
|
ADP-ribosylation factor-like 17B |
| chr4_-_69111401 | 0.51 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr12_+_6419877 | 0.51 |
ENST00000536531.1
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr6_+_36562132 | 0.51 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chrX_-_48824793 | 0.51 |
ENST00000376477.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chrX_-_39923656 | 0.51 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chrX_-_77225135 | 0.50 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr7_+_26331678 | 0.50 |
ENST00000446848.2
|
SNX10
|
sorting nexin 10 |
| chr7_-_142242747 | 0.50 |
ENST00000390362.1
|
TRBV5-3
|
T cell receptor beta variable 5-3 (non-functional) |
| chr1_+_248084320 | 0.50 |
ENST00000319968.4
|
OR2T8
|
olfactory receptor, family 2, subfamily T, member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.7 | 4.3 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.7 | 2.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.7 | 3.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.6 | 1.8 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.5 | 8.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 1.6 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.4 | 1.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.4 | 4.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.4 | 2.8 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.3 | 1.7 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 1.4 | GO:0060584 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.3 | 1.0 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 1.8 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 0.8 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 1.6 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.2 | 5.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 | 1.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 0.9 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 1.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 1.6 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.2 | 0.7 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 0.6 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.2 | 0.8 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.1 | 2.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.2 | GO:0070055 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.1 | 1.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 2.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 3.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.5 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 19.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 2.7 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 2.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.5 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 1.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.8 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.4 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 1.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 14.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.7 | GO:2000767 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 3.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.6 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.4 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 0.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0044053 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 1.9 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.7 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 1.4 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.0 | 1.0 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 1.9 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 1.5 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 3.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 1.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.3 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.4 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.3 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 2.7 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.6 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 1.0 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 2.0 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 2.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 6.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 1.8 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 1.6 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 1.4 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.9 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.7 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 1.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.4 | 7.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 1.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 2.8 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 0.8 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.2 | 0.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 1.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 1.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 2.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 1.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.4 | GO:0043231 | intracellular membrane-bounded organelle(GO:0043231) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 8.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 4.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 2.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.0 | 1.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 4.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 6.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.7 | 2.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.7 | 3.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.6 | 1.8 | GO:0030395 | lactose binding(GO:0030395) |
| 0.5 | 2.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 2.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.3 | 1.6 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 1.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 1.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.9 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.2 | 5.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 5.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.6 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.2 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.8 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 1.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.5 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 2.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 26.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.3 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 1.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 2.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 1.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 2.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 4.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 5.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 5.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 9.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 3.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 3.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 5.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 2.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.2 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |