Project
Illumina Body Map 2, young vs old
Navigation
Downloads
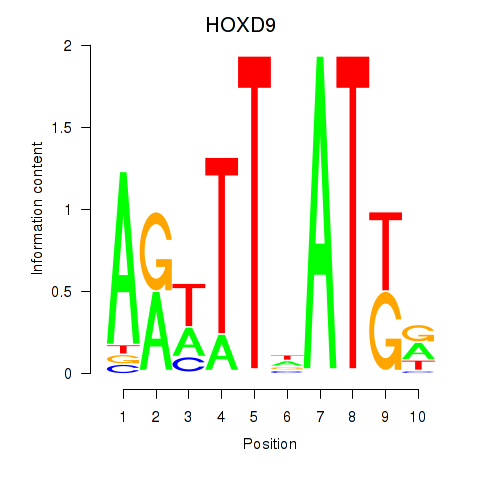
Results for HOXD9
Z-value: 0.09
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.10 | homeobox D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg19_v2_chr2_+_176987088_176987088 | 0.00 | 1.0e+00 | Click! |
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_11463353 | 3.76 |
ENST00000279575.1
ENST00000535904.1 ENST00000445719.2 |
PRB4
|
proline-rich protein BstNI subfamily 4 |
| chr4_-_76944621 | 3.69 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr6_-_52774464 | 3.13 |
ENST00000370968.1
ENST00000211122.3 |
GSTA3
|
glutathione S-transferase alpha 3 |
| chr12_-_11002063 | 2.95 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr11_-_5255861 | 2.59 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr5_+_94727048 | 2.47 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr18_+_61554932 | 2.40 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr4_+_74606223 | 2.34 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr21_+_17792672 | 2.33 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_61151306 | 2.32 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr7_-_16921601 | 2.15 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr2_+_181988620 | 2.04 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr1_+_192127578 | 1.86 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr11_-_104972158 | 1.82 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr6_-_49681235 | 1.80 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr2_+_143635067 | 1.71 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr2_+_89986318 | 1.67 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr2_+_90024732 | 1.63 |
ENST00000390268.2
|
IGKV2D-26
|
immunoglobulin kappa variable 2D-26 |
| chr7_-_38289173 | 1.58 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr8_-_86253888 | 1.55 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr6_-_133055815 | 1.53 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr22_-_30642728 | 1.52 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr17_+_9479944 | 1.49 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr2_-_158345462 | 1.47 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr2_+_90198535 | 1.47 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr1_-_59043166 | 1.46 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr2_-_158345341 | 1.44 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr3_-_167191814 | 1.44 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chrY_+_15815447 | 1.42 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chr15_+_58430368 | 1.41 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr15_+_58430567 | 1.40 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr4_-_76928641 | 1.37 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr1_+_198608292 | 1.34 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr12_+_9980069 | 1.32 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr10_+_115511213 | 1.30 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr5_+_142286887 | 1.29 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr6_-_32634425 | 1.28 |
ENST00000399082.3
ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr8_-_16424871 | 1.26 |
ENST00000518026.1
|
MSR1
|
macrophage scavenger receptor 1 |
| chr14_+_92789498 | 1.24 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr1_+_198608146 | 1.22 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr6_-_49712123 | 1.22 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_+_6588316 | 1.19 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr22_+_22749343 | 1.19 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr12_+_119772502 | 1.19 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr14_-_106725723 | 1.17 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr3_+_178276488 | 1.17 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr17_-_38020379 | 1.15 |
ENST00000351680.3
ENST00000346243.3 ENST00000350532.3 ENST00000467757.1 ENST00000439016.2 |
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr19_-_19739321 | 1.13 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr7_-_121944491 | 1.13 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chrY_+_14958970 | 1.13 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr2_-_89278535 | 1.12 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr12_+_9980113 | 1.12 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr13_-_47012325 | 1.11 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr1_+_198607801 | 1.11 |
ENST00000367379.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr2_+_143635222 | 1.07 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr6_+_32812568 | 1.04 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr10_+_5406935 | 1.03 |
ENST00000380433.3
|
UCN3
|
urocortin 3 |
| chrX_+_36246735 | 1.03 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr3_+_182511266 | 1.01 |
ENST00000323116.5
ENST00000493826.1 |
ATP11B
|
ATPase, class VI, type 11B |
| chr3_-_157221128 | 1.00 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_+_45140360 | 1.00 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr3_-_27498235 | 0.98 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr15_+_49715449 | 0.97 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chrX_+_135730297 | 0.97 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chrX_+_36254051 | 0.95 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chrX_+_36053908 | 0.94 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr12_-_10607084 | 0.94 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr11_-_104905840 | 0.94 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr4_+_71384257 | 0.92 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr3_-_167371704 | 0.90 |
ENST00000488012.1
|
WDR49
|
WD repeat domain 49 |
| chr21_-_31588365 | 0.89 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr7_-_122339162 | 0.87 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr6_-_10115007 | 0.87 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr5_-_39274617 | 0.86 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chr18_+_616672 | 0.85 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr3_+_130162328 | 0.85 |
ENST00000512482.1
|
COL6A5
|
collagen, type VI, alpha 5 |
| chr6_-_47445214 | 0.84 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr6_+_13272904 | 0.83 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr14_+_62164340 | 0.83 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr17_+_54230819 | 0.81 |
ENST00000318698.2
ENST00000566473.2 |
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr7_-_36764004 | 0.79 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr14_+_22615942 | 0.79 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr3_-_116163830 | 0.79 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chrX_+_135730373 | 0.77 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr2_-_188312971 | 0.75 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr19_+_10397648 | 0.74 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr6_+_15401075 | 0.72 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr4_+_39640787 | 0.72 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr3_-_157221380 | 0.72 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr7_+_135710494 | 0.72 |
ENST00000440744.2
|
AC024084.1
|
AC024084.1 |
| chr11_-_104827425 | 0.71 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr6_+_1312675 | 0.71 |
ENST00000296839.2
|
FOXQ1
|
forkhead box Q1 |
| chr18_+_616711 | 0.71 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr1_+_16083098 | 0.71 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr4_+_71296204 | 0.70 |
ENST00000413702.1
|
MUC7
|
mucin 7, secreted |
| chr4_+_169633310 | 0.70 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_+_121465234 | 0.70 |
ENST00000504912.1
ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr7_+_5920429 | 0.70 |
ENST00000242104.5
|
OCM
|
oncomodulin |
| chr19_+_55014013 | 0.70 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr5_-_111093167 | 0.70 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr4_-_68749745 | 0.69 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr11_+_92085262 | 0.69 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr2_+_143886877 | 0.69 |
ENST00000295095.6
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr9_+_12693336 | 0.69 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr1_+_152956549 | 0.69 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr19_+_55385682 | 0.68 |
ENST00000391726.3
|
FCAR
|
Fc fragment of IgA, receptor for |
| chr2_+_114647504 | 0.68 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr2_+_185463093 | 0.67 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr17_-_29645836 | 0.67 |
ENST00000578584.1
|
CTD-2370N5.3
|
CTD-2370N5.3 |
| chr13_-_99959641 | 0.66 |
ENST00000376414.4
|
GPR183
|
G protein-coupled receptor 183 |
| chr4_-_84035868 | 0.65 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr4_-_144940477 | 0.65 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr3_+_111393501 | 0.64 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr4_-_36245561 | 0.64 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr8_-_66701319 | 0.64 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr18_-_24443151 | 0.64 |
ENST00000440832.3
|
AQP4
|
aquaporin 4 |
| chr12_-_91546926 | 0.64 |
ENST00000550758.1
|
DCN
|
decorin |
| chr18_-_5396271 | 0.63 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr14_-_67955426 | 0.63 |
ENST00000554480.1
|
TMEM229B
|
transmembrane protein 229B |
| chr17_+_61086917 | 0.62 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_+_15668240 | 0.62 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr2_+_210518057 | 0.62 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr14_-_68000442 | 0.62 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr1_-_236046872 | 0.61 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr19_+_55014085 | 0.60 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr1_+_82165350 | 0.60 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr12_-_10007448 | 0.60 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr4_-_84035905 | 0.59 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr4_+_75174180 | 0.58 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr21_-_39705323 | 0.58 |
ENST00000436845.1
|
AP001422.3
|
AP001422.3 |
| chr10_-_49459800 | 0.57 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr12_-_95510743 | 0.57 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr14_+_23709555 | 0.57 |
ENST00000430154.2
|
C14orf164
|
chromosome 14 open reading frame 164 |
| chr1_+_172745006 | 0.56 |
ENST00000432694.2
|
RP1-15D23.2
|
RP1-15D23.2 |
| chr4_+_88720698 | 0.56 |
ENST00000226284.5
|
IBSP
|
integrin-binding sialoprotein |
| chr4_+_175839551 | 0.55 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr13_+_33160553 | 0.55 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr1_+_16083123 | 0.54 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr20_-_13971255 | 0.53 |
ENST00000284951.5
ENST00000378072.5 |
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr6_-_131321863 | 0.53 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr15_-_55562582 | 0.53 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_+_107318157 | 0.52 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr11_-_128457446 | 0.52 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr22_+_50609150 | 0.51 |
ENST00000159647.5
ENST00000395842.2 |
PANX2
|
pannexin 2 |
| chr1_+_160370344 | 0.50 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr2_-_163695128 | 0.50 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr9_-_138391692 | 0.50 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chrX_-_125686784 | 0.49 |
ENST00000371126.1
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr10_-_29084886 | 0.49 |
ENST00000608061.1
ENST00000443246.2 ENST00000446012.1 |
LINC00837
|
long intergenic non-protein coding RNA 837 |
| chr6_-_39399087 | 0.49 |
ENST00000229913.5
ENST00000541946.1 ENST00000394362.1 |
KIF6
|
kinesin family member 6 |
| chr12_-_15104040 | 0.49 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_-_26231589 | 0.48 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chr7_+_93535866 | 0.48 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr8_-_25281747 | 0.48 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr11_-_78052923 | 0.48 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr1_+_81771806 | 0.48 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr12_+_14369524 | 0.48 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr14_+_23654525 | 0.47 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr1_-_48937821 | 0.46 |
ENST00000396199.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr14_+_22631122 | 0.46 |
ENST00000390458.3
|
TRAV29DV5
|
T cell receptor alpha variable 29/delta variable 5 (gene/pseudogene) |
| chr5_+_140588269 | 0.46 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr2_+_191334212 | 0.46 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr18_-_53069419 | 0.45 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr14_-_38028689 | 0.45 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr4_+_71384300 | 0.45 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr10_-_105845674 | 0.45 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr9_-_5084580 | 0.45 |
ENST00000601793.1
|
AL161450.1
|
Uncharacterized protein |
| chr2_-_89160117 | 0.45 |
ENST00000390238.2
|
IGKJ5
|
immunoglobulin kappa joining 5 |
| chr7_-_80141328 | 0.44 |
ENST00000398291.3
|
GNAT3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr6_+_130339710 | 0.44 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr14_+_65878650 | 0.44 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr2_+_25015968 | 0.44 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr10_-_99030395 | 0.43 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr4_+_69313145 | 0.43 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr19_-_56826157 | 0.43 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chrX_+_12993336 | 0.42 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr7_+_93535817 | 0.42 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chrX_+_123097014 | 0.42 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr1_-_39339777 | 0.41 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr17_+_57233087 | 0.41 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr17_-_71223839 | 0.41 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr14_-_81425828 | 0.40 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr3_+_140981456 | 0.40 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr17_-_39646116 | 0.40 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr6_-_74231444 | 0.39 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr14_+_57671888 | 0.39 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chrX_-_10645724 | 0.39 |
ENST00000413894.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr2_-_70417827 | 0.39 |
ENST00000457952.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr14_+_23002427 | 0.38 |
ENST00000390527.1
|
TRAJ10
|
T cell receptor alpha joining 10 |
| chr17_+_68071389 | 0.37 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_112564797 | 0.37 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr15_+_66797627 | 0.36 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr13_-_81801115 | 0.36 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr3_+_94657118 | 0.36 |
ENST00000466089.1
ENST00000470465.1 |
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr12_+_9144626 | 0.35 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr10_-_8095412 | 0.35 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr2_-_175712270 | 0.33 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr15_+_52155001 | 0.33 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr9_-_215744 | 0.33 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr1_+_146373546 | 0.33 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.7 | 2.8 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.7 | 2.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.7 | 0.7 | GO:0035700 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.5 | 2.8 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.5 | 1.4 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.4 | 5.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.3 | 1.0 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 1.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 0.7 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.2 | 0.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 1.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.2 | 1.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 0.5 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 0.8 | GO:0070101 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 1.5 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 0.5 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 1.8 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.5 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 1.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 2.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 1.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.7 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.3 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 1.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 2.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.7 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.5 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.3 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 1.7 | GO:0051023 | regulation of immunoglobulin secretion(GO:0051023) |
| 0.1 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 2.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 1.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.2 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 1.3 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 2.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.9 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 5.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.1 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.5 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.9 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.7 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 1.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.7 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 1.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 3.5 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.3 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.0 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.2 | 0.7 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.2 | 0.7 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 0.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 2.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 1.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.3 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.9 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 11.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 5.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.6 | 1.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.6 | 2.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.5 | 1.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.5 | 2.8 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 1.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.3 | 1.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 0.8 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.2 | 1.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 2.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 0.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 0.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 0.7 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 1.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 3.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.5 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.8 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 3.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 2.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 1.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 7.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 5.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 2.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 3.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 7.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 3.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 3.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |