Project
Illumina Body Map 2, young vs old
Navigation
Downloads
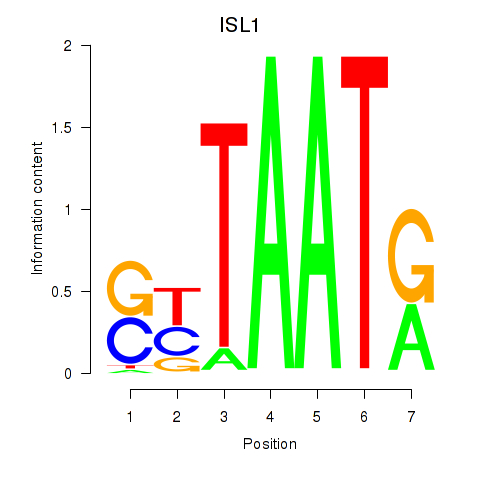
Results for ISL1
Z-value: 0.06
Transcription factors associated with ISL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL1
|
ENSG00000016082.10 | ISL LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL1 | hg19_v2_chr5_+_50678921_50678921 | 0.48 | 5.4e-03 | Click! |
Activity profile of ISL1 motif
Sorted Z-values of ISL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_168155169 | 3.09 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_-_50290839 | 2.89 |
ENST00000552863.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr14_+_62462541 | 2.66 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr14_+_62585332 | 2.52 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr12_-_87232644 | 2.29 |
ENST00000549405.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_-_224467093 | 2.28 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr5_+_140593509 | 2.13 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr1_-_114430169 | 2.13 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr2_-_175711133 | 2.01 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr12_+_79439461 | 1.93 |
ENST00000552624.1
|
SYT1
|
synaptotagmin I |
| chr18_-_40857493 | 1.92 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr9_+_34957477 | 1.89 |
ENST00000544237.1
|
KIAA1045
|
KIAA1045 |
| chr7_-_123673471 | 1.80 |
ENST00000455783.1
|
TMEM229A
|
transmembrane protein 229A |
| chr5_+_137203557 | 1.79 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr2_-_217560248 | 1.75 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr3_-_192445289 | 1.71 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr8_+_70404996 | 1.64 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chrX_-_15619076 | 1.62 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chrX_-_92928557 | 1.60 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr16_-_52119019 | 1.59 |
ENST00000561513.1
ENST00000565742.1 |
LINC00919
|
long intergenic non-protein coding RNA 919 |
| chrX_-_71526999 | 1.58 |
ENST00000453707.2
ENST00000373619.3 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr17_+_67957878 | 1.52 |
ENST00000420427.1
|
AC004562.1
|
AC004562.1 |
| chr5_+_137203465 | 1.51 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr18_+_616672 | 1.50 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr7_-_107880508 | 1.50 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chrX_+_65382433 | 1.50 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr12_+_79357815 | 1.50 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr2_-_224467002 | 1.48 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr2_+_166152283 | 1.45 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_-_176923483 | 1.42 |
ENST00000280187.7
ENST00000512509.1 |
GPM6A
|
glycoprotein M6A |
| chr7_+_103969104 | 1.42 |
ENST00000424859.1
ENST00000535008.1 ENST00000401970.2 ENST00000543266.1 |
LHFPL3
|
lipoma HMGIC fusion partner-like 3 |
| chr3_+_35721106 | 1.42 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr4_+_113970772 | 1.38 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr19_-_22806764 | 1.37 |
ENST00000598042.1
|
AC011516.2
|
AC011516.2 |
| chr4_-_87281196 | 1.37 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr17_+_67957848 | 1.37 |
ENST00000455460.1
|
AC004562.1
|
AC004562.1 |
| chr7_-_81635106 | 1.35 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr4_+_123300121 | 1.33 |
ENST00000446706.1
ENST00000296513.2 |
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr11_-_107729287 | 1.32 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr5_+_169659950 | 1.31 |
ENST00000593851.1
|
C5orf58
|
chromosome 5 open reading frame 58 |
| chr18_+_616711 | 1.31 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr11_-_107729504 | 1.30 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr5_+_137203541 | 1.30 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr4_-_87281224 | 1.29 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr18_+_55018044 | 1.23 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr8_+_35092959 | 1.21 |
ENST00000404895.2
|
UNC5D
|
unc-5 homolog D (C. elegans) |
| chr3_-_107596910 | 1.21 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr9_+_37667978 | 1.20 |
ENST00000539465.1
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr15_+_48498480 | 1.20 |
ENST00000380993.3
ENST00000396577.3 |
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr20_-_5426332 | 1.19 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr11_-_107729887 | 1.18 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_+_207804278 | 1.16 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr7_+_107332334 | 1.16 |
ENST00000541474.1
ENST00000544569.1 |
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr9_+_134165063 | 1.16 |
ENST00000372264.3
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr5_-_67829357 | 1.12 |
ENST00000515199.1
|
CTC-537E7.2
|
CTC-537E7.2 |
| chr8_-_1922789 | 1.11 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr20_+_60174827 | 1.11 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr6_+_42123141 | 1.10 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr8_-_99955042 | 1.09 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr7_-_150675372 | 1.08 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr9_+_103947311 | 1.08 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr2_+_46706725 | 1.07 |
ENST00000434431.1
|
TMEM247
|
transmembrane protein 247 |
| chrX_+_55478538 | 1.06 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr2_-_175711978 | 1.06 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr2_+_173724771 | 1.06 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_-_26744908 | 1.05 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr12_-_28124903 | 1.05 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chrX_-_71526813 | 1.04 |
ENST00000246139.5
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chrX_+_65382381 | 1.02 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr11_-_40315640 | 1.00 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr5_+_173472744 | 1.00 |
ENST00000521585.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr4_+_169418195 | 1.00 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_-_79472365 | 0.98 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr7_-_31380502 | 0.96 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr2_-_68547061 | 0.95 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chrX_-_30327495 | 0.95 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr5_+_140772381 | 0.95 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr5_-_88180342 | 0.94 |
ENST00000502983.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr19_-_50143452 | 0.93 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr16_-_3350614 | 0.92 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr21_-_34185989 | 0.90 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr15_+_59903975 | 0.89 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr11_-_66496430 | 0.89 |
ENST00000533211.1
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2 |
| chr3_+_148508845 | 0.88 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr19_-_17488143 | 0.88 |
ENST00000599426.1
ENST00000252590.4 |
PLVAP
|
plasmalemma vesicle associated protein |
| chr6_+_155537771 | 0.88 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr22_-_32651326 | 0.88 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr9_-_113761720 | 0.86 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chrX_+_30261847 | 0.86 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chrX_-_102941596 | 0.85 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr5_+_140248518 | 0.85 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr7_-_78400364 | 0.85 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_-_42878101 | 0.84 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr22_+_40390930 | 0.84 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chrX_-_49965663 | 0.84 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr16_+_76587314 | 0.84 |
ENST00000563764.1
|
RP11-58C22.1
|
Uncharacterized protein |
| chr1_+_197170592 | 0.83 |
ENST00000535699.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chrX_-_138724994 | 0.83 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr11_-_5531215 | 0.82 |
ENST00000311659.4
|
UBQLN3
|
ubiquilin 3 |
| chr6_+_167704838 | 0.81 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chrX_-_117107680 | 0.81 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr4_+_169418255 | 0.81 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_-_125351481 | 0.80 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr5_-_41794313 | 0.80 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr5_-_102455801 | 0.80 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr4_+_88754113 | 0.79 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr11_-_5531159 | 0.78 |
ENST00000445998.1
|
UBQLN3
|
ubiquilin 3 |
| chr11_-_62783303 | 0.75 |
ENST00000336232.2
ENST00000430500.2 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr20_-_5426380 | 0.74 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr1_-_156217875 | 0.74 |
ENST00000292291.5
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr3_-_100712352 | 0.74 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr17_+_35732916 | 0.74 |
ENST00000586700.1
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chrX_-_117119243 | 0.73 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr13_+_46276441 | 0.72 |
ENST00000310521.1
ENST00000533564.1 |
SPERT
|
spermatid associated |
| chr2_+_172543919 | 0.72 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chrX_-_100129128 | 0.72 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr17_+_35732955 | 0.71 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr11_-_62783276 | 0.71 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr7_-_122342988 | 0.71 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chr1_-_156217822 | 0.71 |
ENST00000368270.1
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr8_-_133637624 | 0.70 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr10_-_116444371 | 0.70 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_+_217804661 | 0.69 |
ENST00000366933.4
|
SPATA17
|
spermatogenesis associated 17 |
| chr6_+_168418553 | 0.69 |
ENST00000354419.2
ENST00000351261.3 |
KIF25
|
kinesin family member 25 |
| chr12_+_96883347 | 0.68 |
ENST00000524981.4
ENST00000298953.3 |
C12orf55
|
chromosome 12 open reading frame 55 |
| chr3_+_35685113 | 0.68 |
ENST00000419330.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_-_149095652 | 0.67 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr21_-_42219065 | 0.67 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr3_-_138312971 | 0.66 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr5_+_32710736 | 0.66 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr18_-_5544241 | 0.66 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr3_+_35721130 | 0.66 |
ENST00000432450.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr11_-_5345582 | 0.66 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr3_-_121740969 | 0.65 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr19_-_50666431 | 0.65 |
ENST00000293405.3
|
IZUMO2
|
IZUMO family member 2 |
| chr18_-_21977748 | 0.65 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr9_-_113100088 | 0.64 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr11_-_64660916 | 0.63 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr8_+_30496078 | 0.63 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr5_+_55354822 | 0.62 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr18_-_22804637 | 0.62 |
ENST00000577775.1
|
ZNF521
|
zinc finger protein 521 |
| chr12_-_5352315 | 0.62 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr13_-_45768841 | 0.60 |
ENST00000379108.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr5_-_138210977 | 0.60 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr2_+_173792893 | 0.59 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_+_32963433 | 0.59 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr20_+_61448376 | 0.58 |
ENST00000343916.3
|
COL9A3
|
collagen, type IX, alpha 3 |
| chr7_+_129932974 | 0.58 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr13_-_70682590 | 0.58 |
ENST00000377844.4
|
KLHL1
|
kelch-like family member 1 |
| chr8_-_94179079 | 0.58 |
ENST00000521906.1
|
C8orf87
|
chromosome 8 open reading frame 87 |
| chr3_-_100551141 | 0.58 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr21_-_34186006 | 0.57 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr2_+_234216454 | 0.57 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr6_-_116575226 | 0.57 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr3_-_100712292 | 0.57 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_-_95391315 | 0.56 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr11_-_102668879 | 0.56 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr17_+_37824217 | 0.56 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chrX_+_114874727 | 0.55 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr14_-_92198403 | 0.55 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr3_-_21792838 | 0.54 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr7_+_138915102 | 0.53 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr11_-_7904464 | 0.53 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr5_+_34757309 | 0.52 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr6_+_167704798 | 0.52 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr12_+_54402790 | 0.52 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr12_+_122356488 | 0.51 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr12_-_6484715 | 0.51 |
ENST00000228916.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr5_-_138211051 | 0.50 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr14_+_21566980 | 0.49 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr10_-_62332357 | 0.49 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_-_186877806 | 0.48 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_37824411 | 0.48 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr1_-_154458520 | 0.47 |
ENST00000486773.1
|
SHE
|
Src homology 2 domain containing E |
| chr6_-_123958141 | 0.47 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr16_+_2198604 | 0.47 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr10_-_75118611 | 0.45 |
ENST00000355577.3
ENST00000394865.1 ENST00000310715.3 ENST00000401621.2 |
TTC18
|
tetratricopeptide repeat domain 18 |
| chr7_-_122342966 | 0.45 |
ENST00000447240.1
|
RNF148
|
ring finger protein 148 |
| chr6_+_31515337 | 0.45 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chrX_-_15683147 | 0.45 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr5_+_102455853 | 0.44 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chrX_+_144908928 | 0.44 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr4_-_9390709 | 0.44 |
ENST00000508324.1
|
RP11-1396O13.13
|
Uncharacterized protein |
| chrX_-_117107542 | 0.44 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr5_-_63258261 | 0.44 |
ENST00000506598.1
|
HTR1A
|
5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled |
| chr10_+_123923205 | 0.43 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_149539053 | 0.43 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr15_+_76352178 | 0.43 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr6_+_90272488 | 0.42 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr3_+_38307293 | 0.42 |
ENST00000311856.4
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr1_+_152178320 | 0.41 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr18_-_47721447 | 0.41 |
ENST00000285039.7
|
MYO5B
|
myosin VB |
| chr12_-_30887948 | 0.40 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chrX_-_23926004 | 0.40 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr12_-_87232771 | 0.39 |
ENST00000550014.1
|
RP11-202H2.1
|
RP11-202H2.1 |
| chr8_+_92261516 | 0.39 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr3_+_157154578 | 0.39 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr2_-_135805008 | 0.38 |
ENST00000414343.1
|
MAP3K19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr1_+_100111479 | 0.38 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr7_+_150811705 | 0.38 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0048174 | negative regulation of short-term neuronal synaptic plasticity(GO:0048174) |
| 0.6 | 1.7 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.6 | 3.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.4 | 3.0 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.4 | 1.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.6 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.3 | 1.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.0 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.2 | 1.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 0.7 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.2 | 0.9 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 0.9 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 3.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 2.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 3.8 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.2 | 1.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.9 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 1.4 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.2 | 0.8 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.2 | 1.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.9 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 3.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.5 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.1 | 0.8 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 1.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.7 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.1 | 1.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.5 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 2.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.1 | 1.1 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.6 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 1.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 1.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.2 | GO:2000625 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.1 | 1.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.3 | GO:0048749 | compound eye development(GO:0048749) |
| 0.1 | 1.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.7 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 3.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.4 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 1.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 2.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.4 | GO:0042407 | inner mitochondrial membrane organization(GO:0007007) cristae formation(GO:0042407) |
| 0.0 | 0.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.9 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.6 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 2.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.9 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 1.8 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 1.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 1.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.9 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 1.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 1.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 1.0 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.6 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.4 | 3.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 1.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 1.8 | GO:0042025 | host cell nucleus(GO:0042025) |
| 0.2 | 3.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.5 | GO:0001534 | radial spoke(GO:0001534) |
| 0.2 | 1.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.0 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 2.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 1.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 1.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 7.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 2.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 4.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 1.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.4 | 1.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 1.0 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.3 | 0.9 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.3 | 2.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 2.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 2.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.7 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 2.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.8 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 3.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 1.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 0.9 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 1.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 3.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 6.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.6 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.9 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 1.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.4 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 3.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 1.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 1.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 2.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.9 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 3.4 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.9 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 4.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 4.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 2.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 4.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |