Project
Illumina Body Map 2, young vs old
Navigation
Downloads
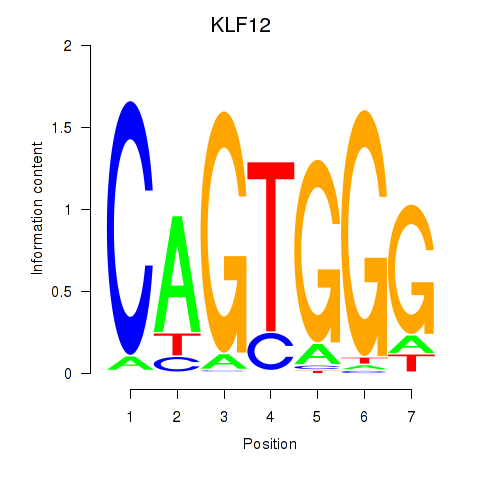
Results for KLF12
Z-value: 0.09
Transcription factors associated with KLF12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF12
|
ENSG00000118922.12 | Kruppel like factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF12 | hg19_v2_chr13_-_74708372_74708409 | -0.19 | 3.0e-01 | Click! |
Activity profile of KLF12 motif
Sorted Z-values of KLF12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_54379569 | 2.18 |
ENST00000513209.1
|
RP11-834C11.12
|
RP11-834C11.12 |
| chr7_-_27224842 | 1.85 |
ENST00000517402.1
|
HOXA11
|
homeobox A11 |
| chr7_-_27224795 | 1.82 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr1_+_153750622 | 1.65 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr11_-_102401469 | 1.48 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr11_-_57089671 | 1.36 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr1_-_201081579 | 1.36 |
ENST00000367338.3
ENST00000362061.3 |
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr12_-_56120838 | 1.34 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr1_+_24118452 | 1.33 |
ENST00000421070.1
|
LYPLA2
|
lysophospholipase II |
| chr16_+_28889801 | 1.31 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr16_+_28889703 | 1.31 |
ENST00000357084.3
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr1_-_43205811 | 1.25 |
ENST00000372539.3
ENST00000296387.1 ENST00000539749.1 |
CLDN19
|
claudin 19 |
| chr5_+_131705597 | 1.22 |
ENST00000435065.2
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr17_+_1665253 | 1.22 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr14_-_23395623 | 1.21 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr11_-_57089774 | 1.21 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr10_-_75401500 | 1.21 |
ENST00000359322.4
|
MYOZ1
|
myozenin 1 |
| chr5_+_131705438 | 1.21 |
ENST00000245407.3
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr4_-_111558135 | 1.21 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chr7_+_143013198 | 1.17 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr6_-_43021612 | 1.14 |
ENST00000535468.1
|
CUL7
|
cullin 7 |
| chr12_+_81101277 | 1.13 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr17_-_80797886 | 1.12 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr10_-_52645416 | 1.10 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr5_-_139930713 | 1.07 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chr17_+_1665306 | 1.05 |
ENST00000571360.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr22_-_24110063 | 1.04 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr4_-_87770416 | 1.04 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr15_-_52944231 | 1.03 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr7_-_135433534 | 1.03 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chr7_+_123295861 | 1.02 |
ENST00000458573.2
ENST00000456238.2 |
LMOD2
|
leiomodin 2 (cardiac) |
| chr7_+_66800928 | 1.02 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr17_+_1665345 | 0.99 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr14_-_61190754 | 0.97 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chrX_+_49028265 | 0.94 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr19_-_3978083 | 0.94 |
ENST00000600794.1
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr2_-_97509749 | 0.93 |
ENST00000331001.2
ENST00000318357.4 |
ANKRD23
|
ankyrin repeat domain 23 |
| chr16_+_85204882 | 0.92 |
ENST00000574293.1
|
CTC-786C10.1
|
CTC-786C10.1 |
| chr1_+_110036699 | 0.91 |
ENST00000496961.1
ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr7_-_100491854 | 0.91 |
ENST00000426415.1
ENST00000430554.1 ENST00000412389.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr12_-_56120865 | 0.91 |
ENST00000548898.1
ENST00000552067.1 |
CD63
|
CD63 molecule |
| chr16_+_89168753 | 0.90 |
ENST00000544543.1
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr6_+_31895467 | 0.89 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr2_-_97509729 | 0.89 |
ENST00000418232.1
|
ANKRD23
|
ankyrin repeat domain 23 |
| chr1_-_19229014 | 0.89 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr10_-_91403625 | 0.87 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr4_-_100575781 | 0.85 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr6_+_133562472 | 0.85 |
ENST00000430974.2
ENST00000367895.5 ENST00000355167.3 ENST00000355286.6 |
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr3_+_148508845 | 0.84 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr3_-_49722523 | 0.84 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr1_+_110036674 | 0.82 |
ENST00000393709.3
|
CYB561D1
|
cytochrome b561 family, member D1 |
| chr9_-_34637806 | 0.82 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr1_-_31661000 | 0.82 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr9_-_131085021 | 0.82 |
ENST00000372890.4
|
TRUB2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr10_+_23983671 | 0.82 |
ENST00000376462.1
|
KIAA1217
|
KIAA1217 |
| chr6_+_133562744 | 0.82 |
ENST00000525849.1
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr1_-_114355083 | 0.82 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr22_-_30198075 | 0.81 |
ENST00000411532.1
|
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chrX_+_153046456 | 0.81 |
ENST00000393786.3
ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3
|
SRSF protein kinase 3 |
| chr3_+_154797428 | 0.80 |
ENST00000460393.1
|
MME
|
membrane metallo-endopeptidase |
| chr9_+_131085095 | 0.80 |
ENST00000372875.3
|
COQ4
|
coenzyme Q4 |
| chr7_+_140373619 | 0.79 |
ENST00000483369.1
|
ADCK2
|
aarF domain containing kinase 2 |
| chr6_+_133562704 | 0.79 |
ENST00000531901.1
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr11_+_117070037 | 0.79 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr16_-_70729496 | 0.78 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr1_+_110036728 | 0.78 |
ENST00000369868.3
ENST00000430195.2 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr19_-_2050852 | 0.77 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr11_-_34533257 | 0.77 |
ENST00000312319.2
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr11_-_64900791 | 0.76 |
ENST00000531018.1
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr6_-_30640811 | 0.75 |
ENST00000376442.3
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr16_-_1843694 | 0.75 |
ENST00000569769.1
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr19_+_6740888 | 0.74 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr19_-_5785630 | 0.74 |
ENST00000586012.1
ENST00000590343.1 |
CTB-54O9.9
DUS3L
|
Uncharacterized protein dihydrouridine synthase 3-like (S. cerevisiae) |
| chr11_-_66103867 | 0.73 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr15_-_75230368 | 0.72 |
ENST00000564811.1
ENST00000562233.1 ENST00000567270.1 ENST00000568783.1 |
COX5A
|
cytochrome c oxidase subunit Va |
| chr16_+_89168993 | 0.72 |
ENST00000538340.1
ENST00000543676.1 |
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr17_+_7210921 | 0.71 |
ENST00000573542.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr5_-_139422654 | 0.70 |
ENST00000289409.4
ENST00000358522.3 ENST00000378238.4 ENST00000289422.7 ENST00000361474.1 ENST00000545385.1 ENST00000394770.1 ENST00000541337.1 |
NRG2
|
neuregulin 2 |
| chrX_+_41583408 | 0.70 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr1_+_214161272 | 0.70 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr2_-_225266743 | 0.69 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr7_-_27702455 | 0.68 |
ENST00000265395.2
|
HIBADH
|
3-hydroxyisobutyrate dehydrogenase |
| chrX_-_129299847 | 0.68 |
ENST00000319908.3
ENST00000287295.3 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr1_+_61330931 | 0.68 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr6_+_31895480 | 0.67 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr2_-_220435963 | 0.66 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr9_-_34637718 | 0.66 |
ENST00000378892.1
ENST00000277010.4 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr9_+_137533615 | 0.66 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chrX_-_129299638 | 0.66 |
ENST00000535724.1
ENST00000346424.2 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr9_-_34590121 | 0.66 |
ENST00000417345.1
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr17_+_1633755 | 0.66 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chr3_-_119379427 | 0.66 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr17_+_7211280 | 0.66 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr10_-_52645379 | 0.65 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr1_+_214161854 | 0.65 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr12_+_53848505 | 0.64 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chrX_+_150866779 | 0.64 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr5_-_179227540 | 0.64 |
ENST00000520875.1
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr16_+_29819096 | 0.64 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_+_172864490 | 0.64 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr19_+_4474846 | 0.62 |
ENST00000589486.1
ENST00000592691.1 |
HDGFRP2
|
Hepatoma-derived growth factor-related protein 2 |
| chr3_+_184037466 | 0.62 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr16_-_67978016 | 0.62 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr14_+_75894714 | 0.61 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr8_+_22022653 | 0.61 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr9_-_34589700 | 0.61 |
ENST00000351266.4
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chrX_+_47082408 | 0.60 |
ENST00000518022.1
ENST00000276052.6 |
CDK16
|
cyclin-dependent kinase 16 |
| chr11_-_2160180 | 0.60 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr22_+_31489344 | 0.60 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr16_+_85646891 | 0.60 |
ENST00000393243.1
|
GSE1
|
Gse1 coiled-coil protein |
| chr8_+_22022800 | 0.60 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr2_-_225266711 | 0.60 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr11_-_64013288 | 0.59 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr17_-_27418537 | 0.59 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr12_+_120740119 | 0.59 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr4_-_185726906 | 0.59 |
ENST00000513317.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr17_-_7155274 | 0.59 |
ENST00000318988.6
ENST00000575783.1 ENST00000573600.1 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr17_+_7210898 | 0.59 |
ENST00000572815.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_-_57328187 | 0.59 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr9_-_34589734 | 0.58 |
ENST00000378980.3
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr16_+_1291240 | 0.58 |
ENST00000561736.1
|
TPSAB1
|
tryptase alpha/beta 1 |
| chr5_-_142077569 | 0.58 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr19_+_50381308 | 0.57 |
ENST00000599049.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr5_-_180632293 | 0.57 |
ENST00000334421.5
|
TRIM7
|
tripartite motif containing 7 |
| chr16_+_69140122 | 0.57 |
ENST00000219322.3
|
HAS3
|
hyaluronan synthase 3 |
| chr19_+_13051206 | 0.57 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr17_-_4889508 | 0.57 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr11_-_63993690 | 0.57 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr7_+_43966018 | 0.57 |
ENST00000222402.3
ENST00000446008.1 ENST00000394798.4 |
UBE2D4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr19_-_46272462 | 0.57 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr19_+_41094612 | 0.57 |
ENST00000595726.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr12_+_53848549 | 0.56 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr17_+_7210852 | 0.56 |
ENST00000576930.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr19_-_46272106 | 0.56 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr3_+_46924829 | 0.56 |
ENST00000313049.5
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr3_-_113415441 | 0.56 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr19_+_10812108 | 0.56 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr5_+_52776449 | 0.56 |
ENST00000396947.3
|
FST
|
follistatin |
| chr1_+_17866290 | 0.56 |
ENST00000361221.3
ENST00000452522.1 ENST00000434513.1 |
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr16_+_85646763 | 0.56 |
ENST00000411612.1
ENST00000253458.7 |
GSE1
|
Gse1 coiled-coil protein |
| chr22_+_47016277 | 0.55 |
ENST00000406902.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr6_-_66417107 | 0.55 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr14_+_63671105 | 0.55 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr14_+_74416989 | 0.55 |
ENST00000334571.2
ENST00000554920.1 |
COQ6
|
coenzyme Q6 monooxygenase |
| chr6_-_30640761 | 0.55 |
ENST00000415603.1
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr11_-_61129306 | 0.55 |
ENST00000544118.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr3_+_29322803 | 0.55 |
ENST00000396583.3
ENST00000383767.2 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr1_-_55341551 | 0.54 |
ENST00000537443.1
|
DHCR24
|
24-dehydrocholesterol reductase |
| chr19_-_46000251 | 0.54 |
ENST00000590526.1
ENST00000344680.4 ENST00000245923.4 |
RTN2
|
reticulon 2 |
| chr19_+_45312347 | 0.54 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr8_+_37553261 | 0.54 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr17_-_2117600 | 0.53 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr9_-_35754253 | 0.53 |
ENST00000436428.2
|
MSMP
|
microseminoprotein, prostate associated |
| chr11_-_62995986 | 0.53 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr1_+_156084461 | 0.53 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr16_+_29818857 | 0.53 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr12_+_121416489 | 0.53 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr6_+_43612750 | 0.52 |
ENST00000372165.4
ENST00000372163.4 |
RSPH9
|
radial spoke head 9 homolog (Chlamydomonas) |
| chr5_+_52776228 | 0.52 |
ENST00000256759.3
|
FST
|
follistatin |
| chr9_-_127952032 | 0.52 |
ENST00000456642.1
ENST00000373546.3 ENST00000373547.4 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr12_-_56122220 | 0.52 |
ENST00000552692.1
|
CD63
|
CD63 molecule |
| chr2_+_128180842 | 0.51 |
ENST00000402125.2
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr6_-_107436473 | 0.50 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr15_-_75230478 | 0.50 |
ENST00000322347.6
|
COX5A
|
cytochrome c oxidase subunit Va |
| chr17_-_40828969 | 0.50 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr12_+_2912363 | 0.50 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr9_-_127952187 | 0.49 |
ENST00000451402.1
ENST00000415905.1 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr12_-_56122124 | 0.49 |
ENST00000552754.1
|
CD63
|
CD63 molecule |
| chr17_-_40829026 | 0.49 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr9_+_131084815 | 0.49 |
ENST00000300452.3
ENST00000609948.1 |
COQ4
|
coenzyme Q4 |
| chr17_-_73266616 | 0.49 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr14_+_75469606 | 0.49 |
ENST00000266126.5
|
EIF2B2
|
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
| chr17_+_78518617 | 0.49 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr5_-_180632147 | 0.49 |
ENST00000274773.7
|
TRIM7
|
tripartite motif containing 7 |
| chr19_+_16338416 | 0.48 |
ENST00000586543.1
|
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr19_-_46389359 | 0.48 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr2_-_74779744 | 0.47 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr16_+_29819372 | 0.47 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr7_+_65552756 | 0.47 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr12_-_56121580 | 0.47 |
ENST00000550776.1
|
CD63
|
CD63 molecule |
| chr1_-_48227290 | 0.47 |
ENST00000594280.1
|
FLJ00388
|
FLJ00388 |
| chr8_-_107782463 | 0.47 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chrX_-_45060135 | 0.46 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr3_+_193853927 | 0.46 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr1_+_159175201 | 0.46 |
ENST00000368121.2
|
DARC
|
Duffy blood group, atypical chemokine receptor |
| chr1_-_203274387 | 0.46 |
ENST00000432511.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr17_+_4981535 | 0.46 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr1_+_32687971 | 0.46 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr19_-_41220957 | 0.46 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr17_-_7154984 | 0.45 |
ENST00000574322.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr2_+_36923830 | 0.45 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr15_-_43865633 | 0.45 |
ENST00000437065.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr14_+_63671577 | 0.45 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr17_-_3499125 | 0.44 |
ENST00000399759.3
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr16_+_29819446 | 0.44 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_+_50380917 | 0.44 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr11_+_66360665 | 0.44 |
ENST00000310190.4
|
CCS
|
copper chaperone for superoxide dismutase |
| chr1_+_155178481 | 0.44 |
ENST00000368376.3
|
MTX1
|
metaxin 1 |
| chrX_-_99986494 | 0.44 |
ENST00000372989.1
ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4
|
synaptotagmin-like 4 |
| chr17_+_41005283 | 0.43 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.8 | 2.4 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.5 | 1.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.5 | 3.3 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.4 | 1.3 | GO:0061114 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.4 | 1.6 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.3 | 2.7 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 3.7 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.3 | 1.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 1.2 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 0.9 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 0.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.7 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.2 | 0.6 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.2 | 2.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 0.6 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.2 | 0.4 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.2 | 0.6 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.2 | 1.9 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.2 | 1.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 1.2 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.2 | 1.8 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.2 | 3.7 | GO:0007501 | mesodermal cell fate specification(GO:0007501) embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.2 | 0.5 | GO:0042668 | inhibition of neuroepithelial cell differentiation(GO:0002085) auditory receptor cell fate determination(GO:0042668) cell-cell signaling involved in cell fate commitment(GO:0045168) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.9 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.4 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.4 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.1 | 0.6 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 1.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.5 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.8 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.5 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.6 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.6 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.8 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.4 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.4 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.1 | 0.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.3 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.1 | 0.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 1.8 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.1 | 1.4 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.6 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 1.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 1.6 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 1.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.5 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 2.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.5 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 1.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.9 | GO:2000767 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.3 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.2 | GO:0032946 | positive regulation of mononuclear cell proliferation(GO:0032946) positive regulation of T cell proliferation(GO:0042102) positive regulation of lymphocyte proliferation(GO:0050671) positive regulation of leukocyte proliferation(GO:0070665) |
| 0.1 | 0.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 3.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.3 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.7 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.6 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 0.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.3 | GO:0072302 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.3 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 1.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.6 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 1.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.3 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.5 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.8 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.7 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 1.2 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.4 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) response to oleic acid(GO:0034201) |
| 0.0 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 1.0 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 1.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.0 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.4 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.5 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.8 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.2 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 1.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.0 | 0.2 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.3 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 1.1 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.8 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.6 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.3 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.9 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.3 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.0 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 | 0.5 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 1.8 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.2 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 0.2 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.3 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 | 0.4 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 1.0 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.4 | 1.8 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 3.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 1.8 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 0.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 2.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 1.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 0.8 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 3.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.0 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.0 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.4 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 2.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.6 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 0.2 | GO:0071745 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 3.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.1 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.3 | 2.4 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.3 | 0.9 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.2 | 2.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 1.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 1.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 0.9 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 0.5 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.2 | 1.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 0.5 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.2 | 0.8 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 1.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 1.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 2.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.6 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.4 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 1.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.3 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 0.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.4 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.3 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 1.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 2.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.4 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 1.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.6 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 1.7 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 3.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 3.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.9 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.0 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.9 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 7.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 3.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 3.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |