Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for KLF15
Z-value: 0.64
Transcription factors associated with KLF15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF15
|
ENSG00000163884.3 | Kruppel like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF15 | hg19_v2_chr3_-_126076264_126076305 | -0.21 | 2.5e-01 | Click! |
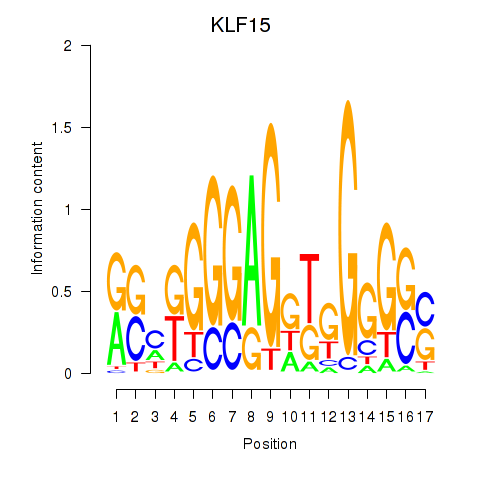
Activity profile of KLF15 motif
Sorted Z-values of KLF15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_47407111 | 2.54 |
ENST00000371904.4
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr11_-_64410787 | 2.53 |
ENST00000301894.2
|
NRXN2
|
neurexin 2 |
| chr20_-_22565101 | 2.37 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr1_-_47407097 | 2.37 |
ENST00000457840.2
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr7_+_29519486 | 2.30 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr19_-_10047219 | 2.11 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr19_+_55795493 | 2.02 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chrX_+_152907913 | 2.01 |
ENST00000370167.4
|
DUSP9
|
dual specificity phosphatase 9 |
| chr14_-_65439132 | 1.97 |
ENST00000533601.2
|
RAB15
|
RAB15, member RAS oncogene family |
| chr16_+_23847339 | 1.90 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr7_+_29519662 | 1.87 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr3_-_27763803 | 1.85 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr8_-_60031762 | 1.84 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr2_-_20425158 | 1.82 |
ENST00000381150.1
|
SDC1
|
syndecan 1 |
| chr5_+_17217669 | 1.81 |
ENST00000322611.3
|
BASP1
|
brain abundant, membrane attached signal protein 1 |
| chr4_+_74702214 | 1.67 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr6_+_31865552 | 1.64 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr7_-_150675372 | 1.59 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr2_-_20424844 | 1.58 |
ENST00000403076.1
ENST00000254351.4 |
SDC1
|
syndecan 1 |
| chr4_-_39640513 | 1.57 |
ENST00000511809.1
ENST00000505729.1 |
SMIM14
|
small integral membrane protein 14 |
| chr19_-_47975106 | 1.55 |
ENST00000539381.1
ENST00000594353.1 ENST00000542837.1 |
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr2_+_20866424 | 1.52 |
ENST00000272224.3
|
GDF7
|
growth differentiation factor 7 |
| chr9_+_130911723 | 1.51 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr7_+_100318423 | 1.47 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr16_+_23847355 | 1.45 |
ENST00000498058.1
|
PRKCB
|
protein kinase C, beta |
| chr14_-_65438865 | 1.43 |
ENST00000267512.5
|
RAB15
|
RAB15, member RAS oncogene family |
| chr2_-_233792837 | 1.43 |
ENST00000373552.4
ENST00000409079.1 |
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr2_-_208030647 | 1.43 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr18_-_74207146 | 1.42 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr3_-_49726104 | 1.41 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr16_+_23847267 | 1.38 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr3_-_27764190 | 1.35 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr11_+_7273181 | 1.34 |
ENST00000318881.6
|
SYT9
|
synaptotagmin IX |
| chr18_+_67068228 | 1.34 |
ENST00000382713.5
|
DOK6
|
docking protein 6 |
| chr12_+_57522692 | 1.33 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr5_-_73937244 | 1.32 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr5_-_111093081 | 1.32 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr17_+_42634844 | 1.31 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr1_-_40367668 | 1.28 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr19_-_47975143 | 1.26 |
ENST00000597014.1
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr9_+_130911770 | 1.25 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr1_-_156051789 | 1.23 |
ENST00000532414.2
|
MEX3A
|
mex-3 RNA binding family member A |
| chr2_-_70780770 | 1.23 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr16_+_56225248 | 1.22 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr20_-_45280066 | 1.18 |
ENST00000279027.4
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr20_-_45280091 | 1.16 |
ENST00000396360.1
ENST00000435032.1 ENST00000413164.2 ENST00000372121.1 ENST00000339636.3 |
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr19_+_11457175 | 1.16 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr14_+_32546274 | 1.15 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr15_-_45480147 | 1.14 |
ENST00000560734.1
|
SHF
|
Src homology 2 domain containing F |
| chr7_-_92463210 | 1.14 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr16_+_2521500 | 1.12 |
ENST00000293973.1
|
NTN3
|
netrin 3 |
| chr19_+_17858509 | 1.11 |
ENST00000594202.1
ENST00000252771.7 ENST00000389133.4 |
FCHO1
|
FCH domain only 1 |
| chr10_+_83634940 | 1.10 |
ENST00000372141.2
ENST00000404547.1 |
NRG3
|
neuregulin 3 |
| chr17_-_42276574 | 1.10 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr17_-_13505219 | 1.09 |
ENST00000284110.1
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr15_-_45480153 | 1.09 |
ENST00000560471.1
ENST00000560540.1 |
SHF
|
Src homology 2 domain containing F |
| chr3_+_11034403 | 1.09 |
ENST00000287766.4
ENST00000425938.1 |
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter), member 1 |
| chr19_+_11457232 | 1.09 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr14_+_90528109 | 1.08 |
ENST00000282146.4
|
KCNK13
|
potassium channel, subfamily K, member 13 |
| chr1_-_32801825 | 1.08 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr19_+_18284477 | 1.08 |
ENST00000407280.3
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr12_+_19282643 | 1.07 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr3_-_49726486 | 1.06 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_-_102323740 | 1.05 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr19_-_11689803 | 1.04 |
ENST00000591319.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr6_-_29595779 | 1.04 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr1_-_177133818 | 1.03 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr15_+_90728145 | 1.02 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr7_+_65670186 | 1.02 |
ENST00000304842.5
ENST00000442120.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr1_-_205313304 | 1.01 |
ENST00000539253.1
ENST00000607826.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr5_+_61602236 | 1.00 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr20_+_13976015 | 0.98 |
ENST00000217246.4
|
MACROD2
|
MACRO domain containing 2 |
| chr2_+_74229812 | 0.98 |
ENST00000305799.7
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr12_+_120933904 | 0.96 |
ENST00000550178.1
ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr2_-_165477971 | 0.96 |
ENST00000446413.2
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr3_-_188665428 | 0.95 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr11_-_46142615 | 0.95 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr14_-_67982146 | 0.95 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr12_+_120933859 | 0.94 |
ENST00000242577.6
ENST00000548214.1 ENST00000392508.2 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr4_-_111120334 | 0.93 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr4_+_74735102 | 0.92 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr18_+_74207477 | 0.91 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr5_+_110559603 | 0.90 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr2_-_73511559 | 0.89 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr5_+_110559784 | 0.89 |
ENST00000282356.4
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr3_+_184097905 | 0.87 |
ENST00000450923.1
|
CHRD
|
chordin |
| chr11_+_22646739 | 0.87 |
ENST00000428556.2
|
AC103801.2
|
AC103801.2 |
| chr4_+_39046615 | 0.87 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr7_+_144052381 | 0.86 |
ENST00000498580.1
ENST00000056217.5 |
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr15_+_57668695 | 0.85 |
ENST00000281282.5
|
CGNL1
|
cingulin-like 1 |
| chr14_-_67981916 | 0.85 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr19_-_1863567 | 0.83 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr2_-_198175495 | 0.83 |
ENST00000409153.1
ENST00000409919.1 ENST00000539527.1 |
ANKRD44
|
ankyrin repeat domain 44 |
| chr17_+_75369400 | 0.83 |
ENST00000590059.1
|
SEPT9
|
septin 9 |
| chr19_-_11689752 | 0.82 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr15_-_38852251 | 0.82 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr4_-_39640700 | 0.81 |
ENST00000295958.5
|
SMIM14
|
small integral membrane protein 14 |
| chr4_-_6474173 | 0.80 |
ENST00000382599.4
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr11_-_46142948 | 0.78 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr1_-_93426998 | 0.77 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr13_+_98795664 | 0.77 |
ENST00000376581.5
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr3_-_10547333 | 0.76 |
ENST00000383800.4
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr7_+_2671568 | 0.76 |
ENST00000258796.7
|
TTYH3
|
tweety family member 3 |
| chr6_-_119670919 | 0.76 |
ENST00000368468.3
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1 |
| chr18_-_5895954 | 0.76 |
ENST00000581347.2
|
TMEM200C
|
transmembrane protein 200C |
| chr3_+_101546827 | 0.75 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr3_+_184098065 | 0.74 |
ENST00000348986.3
|
CHRD
|
chordin |
| chr14_+_92980111 | 0.73 |
ENST00000216487.7
ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr1_-_241520385 | 0.72 |
ENST00000366564.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr3_+_195943369 | 0.71 |
ENST00000296327.5
|
SLC51A
|
solute carrier family 51, alpha subunit |
| chr4_+_79697495 | 0.71 |
ENST00000502871.1
ENST00000335016.5 |
BMP2K
|
BMP2 inducible kinase |
| chr2_-_61697862 | 0.70 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr3_+_184097836 | 0.70 |
ENST00000204604.1
ENST00000310236.3 |
CHRD
|
chordin |
| chr11_-_72496976 | 0.70 |
ENST00000539138.1
ENST00000542989.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr5_-_11904152 | 0.70 |
ENST00000304623.8
ENST00000458100.2 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr22_+_48972118 | 0.69 |
ENST00000358295.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr11_-_71159458 | 0.69 |
ENST00000355527.3
|
DHCR7
|
7-dehydrocholesterol reductase |
| chr5_-_11904100 | 0.68 |
ENST00000359640.2
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr7_-_142120321 | 0.68 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chr4_-_170947446 | 0.68 |
ENST00000507601.1
ENST00000512698.1 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr7_-_92465868 | 0.68 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr7_-_767249 | 0.68 |
ENST00000403562.1
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr3_-_10547192 | 0.67 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr22_+_42949925 | 0.67 |
ENST00000327678.5
ENST00000340239.4 ENST00000407614.4 ENST00000335879.5 |
SERHL2
|
serine hydrolase-like 2 |
| chr15_-_27018175 | 0.67 |
ENST00000311550.5
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr3_+_46923670 | 0.67 |
ENST00000427125.2
ENST00000430002.2 |
PTH1R
|
parathyroid hormone 1 receptor |
| chr6_+_6588316 | 0.67 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr18_+_13218769 | 0.67 |
ENST00000399848.3
ENST00000361205.4 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr10_+_35415851 | 0.66 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr5_-_141257954 | 0.66 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr9_+_101867387 | 0.66 |
ENST00000374990.2
ENST00000552516.1 |
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr9_+_101867359 | 0.66 |
ENST00000374994.4
|
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr6_-_143832793 | 0.65 |
ENST00000438118.2
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr10_+_21823079 | 0.64 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_-_38273840 | 0.64 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr17_+_30593195 | 0.64 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr1_-_151689259 | 0.64 |
ENST00000420342.1
ENST00000290583.4 |
CELF3
|
CUGBP, Elav-like family member 3 |
| chr19_-_42636617 | 0.64 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr1_+_173837488 | 0.64 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr22_+_22673051 | 0.64 |
ENST00000390289.2
|
IGLV5-52
|
immunoglobulin lambda variable 5-52 |
| chr6_-_32157947 | 0.63 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr7_-_105029329 | 0.63 |
ENST00000393651.3
ENST00000460391.1 |
SRPK2
|
SRSF protein kinase 2 |
| chr7_-_28220354 | 0.63 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr1_-_241520525 | 0.63 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr4_+_174089951 | 0.62 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr13_-_52026730 | 0.62 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr11_+_17756279 | 0.62 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr11_+_369804 | 0.62 |
ENST00000329962.6
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr4_-_170947485 | 0.62 |
ENST00000504999.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr2_+_5832799 | 0.61 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr2_-_220173685 | 0.61 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr19_+_589893 | 0.60 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr13_+_95364963 | 0.60 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr22_-_43583079 | 0.60 |
ENST00000216129.6
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr2_+_39893043 | 0.59 |
ENST00000281961.2
|
TMEM178A
|
transmembrane protein 178A |
| chr6_-_119670898 | 0.59 |
ENST00000368466.2
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1 |
| chr21_-_40720995 | 0.59 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr5_+_61601965 | 0.59 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr11_-_8954491 | 0.59 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr16_+_81478775 | 0.58 |
ENST00000537098.3
|
CMIP
|
c-Maf inducing protein |
| chr7_+_94285637 | 0.58 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr2_-_9143786 | 0.58 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr2_+_27371866 | 0.58 |
ENST00000296096.5
|
TCF23
|
transcription factor 23 |
| chr17_-_42277203 | 0.57 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr5_+_139027877 | 0.57 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr15_-_72766533 | 0.56 |
ENST00000562573.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr7_+_30323923 | 0.56 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
| chr11_-_64612041 | 0.56 |
ENST00000342711.5
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chr6_-_143832820 | 0.55 |
ENST00000002165.6
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr22_-_50970506 | 0.55 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr6_+_24495185 | 0.55 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr13_-_49107303 | 0.55 |
ENST00000344532.3
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr16_-_57513657 | 0.54 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr11_+_45918092 | 0.54 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr20_-_48099182 | 0.53 |
ENST00000371741.4
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr15_-_34875771 | 0.53 |
ENST00000267731.7
|
GOLGA8B
|
golgin A8 family, member B |
| chr14_+_32546485 | 0.53 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr7_-_94285511 | 0.53 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr19_-_11688951 | 0.53 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr17_+_65821780 | 0.52 |
ENST00000321892.4
ENST00000335221.5 ENST00000306378.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr2_-_9771075 | 0.51 |
ENST00000446619.1
ENST00000238081.3 |
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr2_-_165697920 | 0.51 |
ENST00000342193.4
ENST00000375458.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr20_-_62710832 | 0.51 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19 |
| chr16_+_50187556 | 0.50 |
ENST00000561678.1
ENST00000357464.3 |
PAPD5
|
PAP associated domain containing 5 |
| chr9_-_6015607 | 0.50 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr19_-_47975417 | 0.50 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr10_+_114709999 | 0.50 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_+_238273 | 0.50 |
ENST00000256509.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr12_+_56435637 | 0.50 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr3_+_184279566 | 0.50 |
ENST00000330394.2
|
EPHB3
|
EPH receptor B3 |
| chr6_+_31683117 | 0.50 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr16_+_50300427 | 0.50 |
ENST00000394697.2
ENST00000566433.2 ENST00000538642.1 |
ADCY7
|
adenylate cyclase 7 |
| chr3_-_197282821 | 0.50 |
ENST00000445160.2
ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr1_+_206730484 | 0.49 |
ENST00000304534.8
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr11_+_117049854 | 0.49 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr5_+_61602055 | 0.49 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr12_+_123849462 | 0.49 |
ENST00000543072.1
|
hsa-mir-8072
|
hsa-mir-8072 |
| chr1_+_180199393 | 0.49 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr13_-_52027134 | 0.48 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr15_-_65067773 | 0.48 |
ENST00000300069.4
|
RBPMS2
|
RNA binding protein with multiple splicing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 1.1 | 3.4 | GO:0048627 | myoblast development(GO:0048627) |
| 1.1 | 3.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.0 | 4.9 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.8 | 2.3 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.6 | 1.8 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.6 | 2.4 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.6 | 2.8 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.5 | 1.6 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 2.3 | GO:0015744 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) succinate transmembrane transport(GO:0071422) |
| 0.4 | 1.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.4 | 1.8 | GO:2001076 | regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.3 | 1.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 1.0 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.3 | 2.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 0.8 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.3 | 2.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 1.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.3 | 1.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.3 | 1.3 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.2 | 1.5 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.9 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 0.4 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.2 | 1.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 1.3 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.2 | 1.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 2.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.3 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.2 | 0.7 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.2 | 0.5 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 0.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.5 | GO:0050894 | determination of affect(GO:0050894) |
| 0.2 | 0.5 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.4 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.9 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 3.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.0 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 2.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 2.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 1.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.8 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.7 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.3 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 2.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 1.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.4 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 2.6 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.6 | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway(GO:0090101) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.6 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.5 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 1.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.7 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.1 | GO:0072054 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 | 0.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.3 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 1.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.9 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.7 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:1900390 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.2 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.1 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.1 | 0.2 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.1 | 0.4 | GO:1901490 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) regulation of lymphangiogenesis(GO:1901490) |
| 0.1 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 1.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.3 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.5 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.1 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.7 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 1.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.4 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.6 | GO:0035864 | response to potassium ion(GO:0035864) |
| 0.0 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 3.7 | GO:1903307 | positive regulation of regulated secretory pathway(GO:1903307) |
| 0.0 | 1.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.7 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.7 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.5 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.4 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 1.8 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.9 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 1.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 1.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.2 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.7 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 2.1 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.5 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 1.0 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.5 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.5 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.7 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.6 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.6 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.5 | GO:0048557 | embryonic digestive tract morphogenesis(GO:0048557) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:1990418 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.5 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.4 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 1.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 0.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 3.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 2.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 4.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 7.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.0 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 4.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.5 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.0 | 4.9 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.6 | 1.8 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 1.5 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.4 | 1.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.4 | 1.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.4 | 2.3 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.3 | 1.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.3 | 0.9 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.2 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 2.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.6 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.2 | 1.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 1.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 1.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 3.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.5 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.2 | 2.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 2.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.3 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 0.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.4 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 2.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.5 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 1.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.3 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 1.3 | GO:0005114 | transforming growth factor beta receptor activity, type I(GO:0005025) type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.4 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.3 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 1.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.6 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.5 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 2.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 7.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 3.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 2.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 2.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 2.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 1.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 2.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.0 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 1.1 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 4.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 3.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 2.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 4.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 4.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 3.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 3.8 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 2.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.6 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 3.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |