Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for KLF7
Z-value: 0.13
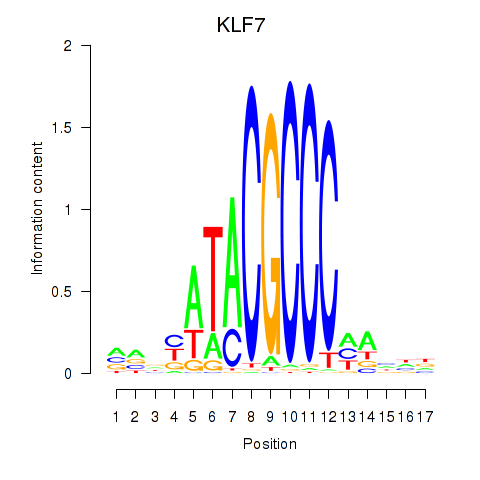
Transcription factors associated with KLF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF7
|
ENSG00000118263.10 | Kruppel like factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF7 | hg19_v2_chr2_-_208031943_208031991 | -0.49 | 4.4e-03 | Click! |
Activity profile of KLF7 motif
Sorted Z-values of KLF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_186353756 | 2.33 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr1_-_19229014 | 2.08 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr1_-_19229248 | 2.01 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr1_-_19229218 | 1.82 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr16_+_20499024 | 1.67 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr6_+_31865552 | 1.58 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr1_-_21948906 | 1.46 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr8_-_6837602 | 1.44 |
ENST00000382692.2
|
DEFA1
|
defensin, alpha 1 |
| chr8_-_6875778 | 1.35 |
ENST00000535841.1
ENST00000327857.2 |
DEFA1B
DEFA3
|
defensin, alpha 1B defensin, alpha 3, neutrophil-specific |
| chr11_+_18417948 | 0.78 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr15_+_96873921 | 0.77 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_188430478 | 0.62 |
ENST00000421427.1
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr16_+_31539183 | 0.61 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr16_-_65268795 | 0.61 |
ENST00000565901.1
ENST00000567058.1 |
RP11-256I9.2
|
RP11-256I9.2 |
| chr14_+_38660185 | 0.54 |
ENST00000555655.1
|
CTD-2142D14.1
|
CTD-2142D14.1 |
| chr18_+_72166564 | 0.54 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr17_+_41476327 | 0.52 |
ENST00000320033.4
|
ARL4D
|
ADP-ribosylation factor-like 4D |
| chr6_+_30585486 | 0.51 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr8_+_115294296 | 0.44 |
ENST00000521523.1
ENST00000523682.1 ENST00000507929.2 ENST00000520543.1 ENST00000519248.1 |
RP11-267L5.1
|
RP11-267L5.1 |
| chr3_+_105085734 | 0.40 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr3_-_178865747 | 0.34 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr16_+_31539197 | 0.33 |
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr13_-_108867101 | 0.32 |
ENST00000356922.4
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr1_-_6453426 | 0.32 |
ENST00000545482.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr1_-_6453399 | 0.30 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr12_-_49581152 | 0.29 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr7_-_47492782 | 0.29 |
ENST00000413551.1
|
TNS3
|
tensin 3 |
| chr19_+_39390587 | 0.24 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr17_+_73008755 | 0.23 |
ENST00000584208.1
ENST00000301585.5 |
ICT1
|
immature colon carcinoma transcript 1 |
| chr3_-_33138624 | 0.22 |
ENST00000445488.2
ENST00000307377.8 ENST00000440656.1 ENST00000436768.1 ENST00000307363.5 |
GLB1
|
galactosidase, beta 1 |
| chr6_-_109703634 | 0.22 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chr5_+_141348598 | 0.21 |
ENST00000394520.2
ENST00000347642.3 |
RNF14
|
ring finger protein 14 |
| chr19_-_14629224 | 0.20 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr9_+_130565147 | 0.19 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr8_+_72587535 | 0.19 |
ENST00000519840.1
ENST00000521131.1 |
RP11-1144P22.1
|
RP11-1144P22.1 |
| chr5_+_141348721 | 0.19 |
ENST00000507163.1
ENST00000394519.1 |
RNF14
|
ring finger protein 14 |
| chr2_+_74710194 | 0.18 |
ENST00000410003.1
ENST00000442235.2 ENST00000233623.5 |
TTC31
|
tetratricopeptide repeat domain 31 |
| chr6_-_33239612 | 0.17 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr19_+_7694623 | 0.16 |
ENST00000594797.1
ENST00000456958.3 ENST00000601406.1 |
PET100
|
PET100 homolog (S. cerevisiae) |
| chr6_-_109703600 | 0.16 |
ENST00000512821.1
|
CD164
|
CD164 molecule, sialomucin |
| chr6_-_33239712 | 0.16 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr6_-_109703663 | 0.15 |
ENST00000368961.5
|
CD164
|
CD164 molecule, sialomucin |
| chr9_+_130565568 | 0.12 |
ENST00000423577.1
|
FPGS
|
folylpolyglutamate synthase |
| chr6_+_43603552 | 0.12 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr19_-_40931891 | 0.11 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr16_+_81772633 | 0.11 |
ENST00000566191.1
ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1
PLCG2
|
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr1_+_87380299 | 0.10 |
ENST00000370551.4
ENST00000370550.5 |
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr5_+_173315283 | 0.10 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr19_+_39390320 | 0.10 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr22_-_30987837 | 0.09 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr9_+_130565487 | 0.08 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chr12_-_49582593 | 0.06 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr3_+_183817967 | 0.06 |
ENST00000335304.2
ENST00000431041.1 ENST00000436361.2 ENST00000440596.2 |
HTR3E
|
5-hydroxytryptamine (serotonin) receptor 3E, ionotropic |
| chr8_+_17104539 | 0.05 |
ENST00000521829.1
ENST00000521005.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr3_+_178866199 | 0.04 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr22_-_30987849 | 0.04 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr2_-_61765732 | 0.03 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr19_-_39390350 | 0.03 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr17_-_18218270 | 0.03 |
ENST00000321105.5
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr15_+_26147507 | 0.02 |
ENST00000383019.2
ENST00000557171.1 |
RP11-1084I9.1
|
RP11-1084I9.1 |
| chr17_-_48277552 | 0.02 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chrX_+_102631844 | 0.02 |
ENST00000372634.1
ENST00000299872.7 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr19_+_58011258 | 0.02 |
ENST00000593916.1
ENST00000598770.1 ENST00000599847.1 ENST00000282292.4 |
ZNF773
|
zinc finger protein 773 |
| chr1_-_21503337 | 0.02 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr2_+_67624430 | 0.00 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr2_-_74710078 | 0.00 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chr6_-_30585009 | 0.00 |
ENST00000376511.2
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.9 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.5 | 1.5 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.4 | 1.4 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.2 | 0.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.6 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.4 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 0.3 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.3 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 1.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 1.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.2 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.0 | 2.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.9 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 3.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.2 | 0.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.4 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.1 | 0.5 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 2.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 6.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |