Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for LMX1B_MNX1_RAX2
Z-value: 0.21
Transcription factors associated with LMX1B_MNX1_RAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
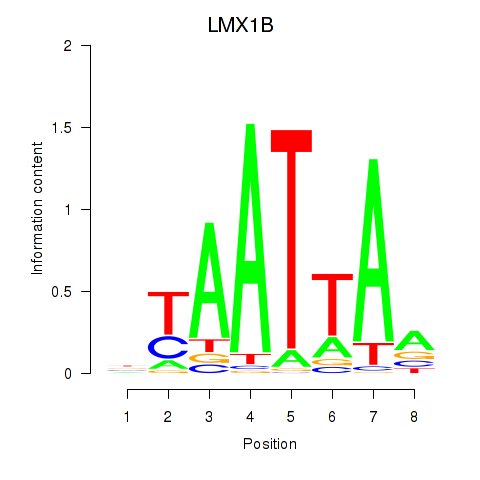
LMX1B
|
ENSG00000136944.13 | LIM homeobox transcription factor 1 beta |
|
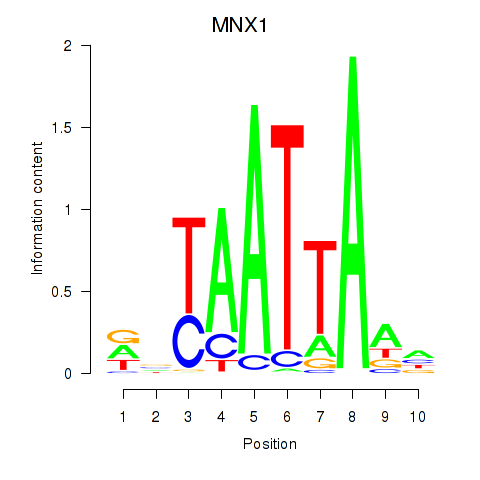
MNX1
|
ENSG00000130675.10 | motor neuron and pancreas homeobox 1 |
|
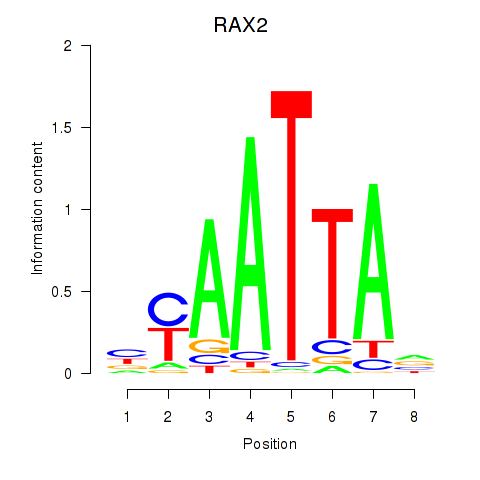
RAX2
|
ENSG00000173976.11 | retina and anterior neural fold homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MNX1 | hg19_v2_chr7_-_156803329_156803362 | 0.66 | 4.4e-05 | Click! |
| LMX1B | hg19_v2_chr9_+_129376722_129376748 | 0.53 | 1.7e-03 | Click! |
| RAX2 | hg19_v2_chr19_-_3772209_3772236 | 0.19 | 3.1e-01 | Click! |
Activity profile of LMX1B_MNX1_RAX2 motif
Sorted Z-values of LMX1B_MNX1_RAX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_24298531 | 3.31 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr8_+_24298597 | 2.95 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr15_+_48483736 | 2.65 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr3_+_173116225 | 2.50 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr7_-_92855762 | 2.43 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr4_+_169418195 | 2.08 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_41831485 | 2.07 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr4_-_138453606 | 2.05 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr17_-_46690839 | 2.04 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr16_-_29910853 | 2.01 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr18_+_76740189 | 1.88 |
ENST00000537592.2
ENST00000575389.2 |
SALL3
|
spalt-like transcription factor 3 |
| chr8_+_24298438 | 1.85 |
ENST00000441335.2
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr5_+_174151536 | 1.79 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr1_-_234667504 | 1.70 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chrX_+_99839799 | 1.49 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr6_-_87804815 | 1.49 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr3_-_151034734 | 1.47 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr7_-_107443652 | 1.46 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr8_+_50824233 | 1.45 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr6_+_78400375 | 1.43 |
ENST00000602452.2
|
MEI4
|
meiosis-specific 4 homolog (S. cerevisiae) |
| chr4_-_138453559 | 1.36 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chrY_-_13524717 | 1.34 |
ENST00000331172.6
|
SLC9B1P1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 pseudogene 1 |
| chr15_-_37393406 | 1.30 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr4_+_169418255 | 1.28 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_56833184 | 1.27 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr1_-_101360331 | 1.26 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr5_-_24645078 | 1.20 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr1_-_101360205 | 1.12 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr1_-_190446759 | 1.11 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr13_+_53602894 | 1.09 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chrX_-_10851762 | 1.08 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr8_+_105235572 | 1.08 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr15_+_76352178 | 1.07 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr5_+_176811431 | 1.07 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr9_+_12693336 | 1.05 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr8_-_109799793 | 1.04 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr12_-_10978957 | 1.03 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr7_+_134464376 | 1.02 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr9_-_95166841 | 1.02 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chrX_-_13835147 | 1.01 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr12_+_26348246 | 1.01 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr7_-_130080977 | 0.99 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr9_-_95166884 | 0.98 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr1_-_101360374 | 0.96 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr1_+_152943122 | 0.96 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr18_+_32173276 | 0.94 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr4_+_114214125 | 0.94 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr4_-_20985632 | 0.92 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr13_-_36788718 | 0.92 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr11_-_26744908 | 0.92 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr1_-_242612779 | 0.91 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr11_-_129062093 | 0.91 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr20_-_29978383 | 0.91 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr1_+_160370344 | 0.89 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr1_-_185597619 | 0.87 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr5_+_31193847 | 0.86 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr13_-_86373536 | 0.85 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr5_+_126984710 | 0.84 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr4_+_105828537 | 0.82 |
ENST00000515649.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr4_+_71108300 | 0.81 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr4_+_105828492 | 0.81 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr3_-_12587055 | 0.81 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr2_+_3800169 | 0.80 |
ENST00000399143.3
|
DCDC2C
|
doublecortin domain containing 2C |
| chr1_-_204135450 | 0.79 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr3_+_111717511 | 0.78 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr8_-_1922789 | 0.78 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr12_+_26348429 | 0.77 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr20_-_56647116 | 0.75 |
ENST00000441277.2
ENST00000452842.1 |
RP13-379L11.1
|
RP13-379L11.1 |
| chr17_-_38859996 | 0.75 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr12_-_6233828 | 0.75 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr20_-_29978286 | 0.75 |
ENST00000376315.2
|
DEFB119
|
defensin, beta 119 |
| chr2_+_149974684 | 0.74 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr18_+_34124507 | 0.74 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr1_-_94586651 | 0.73 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr6_-_31107127 | 0.71 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr11_-_16419067 | 0.67 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr4_+_69313145 | 0.66 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr1_+_160160283 | 0.66 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_-_149293990 | 0.64 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr7_+_134464414 | 0.64 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr7_-_107642348 | 0.63 |
ENST00000393561.1
|
LAMB1
|
laminin, beta 1 |
| chr12_-_94673956 | 0.63 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr2_+_179318295 | 0.61 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr11_-_102576537 | 0.61 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr12_+_26348582 | 0.61 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr10_+_45406627 | 0.61 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr5_-_147286065 | 0.61 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr1_+_160160346 | 0.59 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_+_111717600 | 0.59 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr5_-_36301984 | 0.59 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chrX_-_139047669 | 0.57 |
ENST00000370540.1
|
CXorf66
|
chromosome X open reading frame 66 |
| chr1_+_107683436 | 0.56 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr1_-_95783809 | 0.56 |
ENST00000423410.1
|
RP4-586O15.1
|
RP4-586O15.1 |
| chr6_-_100912785 | 0.56 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr1_+_107682629 | 0.55 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr9_-_95166976 | 0.55 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr3_+_111718036 | 0.54 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr2_+_177015122 | 0.54 |
ENST00000468418.3
|
HOXD3
|
homeobox D3 |
| chr15_+_69857515 | 0.53 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr12_-_52967600 | 0.53 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr1_-_242612726 | 0.52 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr8_-_139926236 | 0.52 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
| chr4_-_87028478 | 0.51 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_-_112123524 | 0.50 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr22_-_28490123 | 0.50 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr2_+_78143006 | 0.48 |
ENST00000443419.1
|
AC073628.1
|
AC073628.1 |
| chr10_+_24497704 | 0.47 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr14_+_65016620 | 0.47 |
ENST00000298705.1
|
PPP1R36
|
protein phosphatase 1, regulatory subunit 36 |
| chr6_+_105404899 | 0.46 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr12_-_91573132 | 0.44 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr7_-_83278322 | 0.44 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr10_-_75351088 | 0.44 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr16_-_14109841 | 0.44 |
ENST00000576797.1
ENST00000575424.1 |
CTD-2135D7.5
|
CTD-2135D7.5 |
| chr13_-_44735393 | 0.43 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr12_-_91573249 | 0.43 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr14_-_21058982 | 0.43 |
ENST00000556526.1
|
RNASE12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr1_+_115572415 | 0.43 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr4_-_186696425 | 0.43 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_+_23709555 | 0.43 |
ENST00000430154.2
|
C14orf164
|
chromosome 14 open reading frame 164 |
| chr21_-_42219065 | 0.43 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr21_+_35736302 | 0.43 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr10_+_115511434 | 0.42 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr2_-_50574856 | 0.42 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr5_+_61874562 | 0.40 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr21_+_17442799 | 0.40 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_+_54410664 | 0.39 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr6_-_167797887 | 0.39 |
ENST00000476779.2
ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10
|
t-complex 10 |
| chr8_+_117950422 | 0.39 |
ENST00000378279.3
|
AARD
|
alanine and arginine rich domain containing protein |
| chr5_+_166711804 | 0.39 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr19_-_14064114 | 0.39 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr9_+_12695702 | 0.39 |
ENST00000381136.2
|
TYRP1
|
tyrosinase-related protein 1 |
| chr4_+_147145709 | 0.38 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr13_+_34922173 | 0.38 |
ENST00000605909.1
|
RP11-16D22.2
|
RP11-16D22.2 |
| chr3_-_157221128 | 0.38 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr5_+_61874696 | 0.38 |
ENST00000491184.2
|
LRRC70
|
leucine rich repeat containing 70 |
| chr4_+_88571429 | 0.38 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr10_+_115511213 | 0.37 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr7_+_129984630 | 0.37 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr3_-_79816965 | 0.37 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr7_-_84121858 | 0.36 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr19_-_7697857 | 0.36 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr5_-_126409159 | 0.36 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr3_+_115342349 | 0.36 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr9_-_28670283 | 0.36 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr6_-_76072719 | 0.35 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr10_+_32873190 | 0.35 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr3_-_20053741 | 0.35 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr18_+_5748793 | 0.35 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chr6_+_153071925 | 0.35 |
ENST00000367244.3
ENST00000367243.3 |
VIP
|
vasoactive intestinal peptide |
| chr10_+_118083919 | 0.35 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr11_-_124190184 | 0.34 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr17_+_72427477 | 0.34 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr3_-_164796269 | 0.33 |
ENST00000264382.3
|
SI
|
sucrase-isomaltase (alpha-glucosidase) |
| chr2_-_180610767 | 0.33 |
ENST00000409343.1
|
ZNF385B
|
zinc finger protein 385B |
| chr1_+_40713573 | 0.33 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr9_+_130026756 | 0.33 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr1_+_107683644 | 0.32 |
ENST00000370067.1
|
NTNG1
|
netrin G1 |
| chr7_-_99716940 | 0.32 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_+_187454749 | 0.32 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr6_+_4087664 | 0.31 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr6_-_49937338 | 0.31 |
ENST00000398718.1
|
DEFB113
|
defensin, beta 113 |
| chr4_+_169633310 | 0.31 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_+_97361388 | 0.31 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr18_+_68002675 | 0.31 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr12_+_78359999 | 0.30 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr14_+_32798547 | 0.30 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_68150744 | 0.30 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr14_+_32798462 | 0.30 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr18_-_33709268 | 0.30 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr2_-_190927447 | 0.30 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr5_+_53751445 | 0.30 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr1_-_67266939 | 0.30 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr7_-_111032971 | 0.30 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr19_-_56110859 | 0.29 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr1_-_186430222 | 0.29 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr11_-_4719072 | 0.29 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr9_-_98189055 | 0.29 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr1_-_227505289 | 0.29 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr12_-_16759440 | 0.28 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_-_86650077 | 0.28 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chrM_+_10053 | 0.28 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr19_-_7698599 | 0.28 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr11_-_30608413 | 0.28 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr17_+_42785976 | 0.28 |
ENST00000393547.2
ENST00000398338.3 |
DBF4B
|
DBF4 homolog B (S. cerevisiae) |
| chr1_+_50569575 | 0.27 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr2_+_171034646 | 0.27 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr6_+_12290586 | 0.27 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr20_-_50722183 | 0.26 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr7_-_99716952 | 0.26 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_46716647 | 0.26 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chrM_+_7586 | 0.26 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr5_-_1882858 | 0.26 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr4_+_3388057 | 0.26 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr7_-_99717463 | 0.26 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_+_155538093 | 0.25 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_+_167584081 | 0.25 |
ENST00000366832.2
|
TCP10L2
|
t-complex 10-like 2 |
| chr18_-_500692 | 0.25 |
ENST00000400256.3
|
COLEC12
|
collectin sub-family member 12 |
| chr9_-_16728161 | 0.25 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr9_+_44867571 | 0.25 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr12_-_23737534 | 0.24 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LMX1B_MNX1_RAX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.6 | 1.8 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.5 | 1.4 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.4 | 0.4 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.4 | 1.1 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.3 | 0.9 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.3 | 3.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 1.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 1.5 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 3.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.8 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 0.6 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.7 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 2.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 2.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 1.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 0.7 | GO:0072378 | blood coagulation, intrinsic pathway(GO:0007597) blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.3 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.0 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 1.5 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 1.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.3 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.3 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.3 | GO:0060585 | nitric oxide transport(GO:0030185) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.9 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 2.5 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.3 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 1.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.3 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.4 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.6 | GO:0072257 | negative regulation of catenin import into nucleus(GO:0035414) metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.3 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 5.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.1 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 1.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 2.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 2.8 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 1.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 2.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.9 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 0.6 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.2 | 1.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 1.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0034686 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 2.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 2.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 3.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.9 | 2.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.4 | 1.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 3.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.0 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.4 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 8.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 1.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 4.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |