Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for MAFF_MAFG
Z-value: 0.39
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
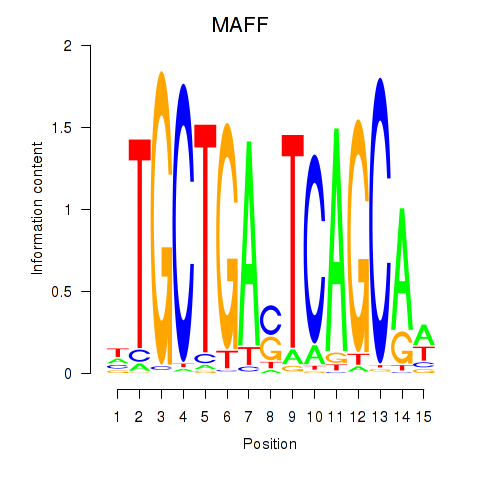
MAFF
|
ENSG00000185022.7 | MAF bZIP transcription factor F |
|
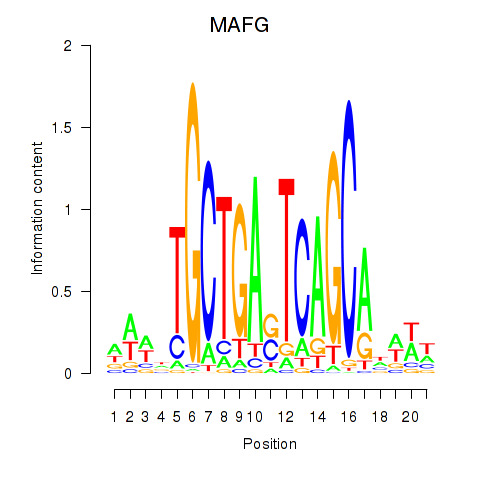
MAFG
|
ENSG00000197063.6 | MAF bZIP transcription factor G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFG | hg19_v2_chr17_-_79881408_79881423 | -0.23 | 2.1e-01 | Click! |
| MAFF | hg19_v2_chr22_+_38609538_38609547 | 0.08 | 6.5e-01 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_2709527 | 6.29 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr22_+_21133469 | 3.51 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr12_+_20968608 | 2.55 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chrY_+_2709906 | 2.48 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr3_+_186383741 | 2.25 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr11_+_75428857 | 2.12 |
ENST00000198801.5
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2 |
| chr2_+_211421262 | 1.92 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chrX_+_138612889 | 1.64 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr1_+_196912902 | 1.55 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr2_+_128177253 | 1.50 |
ENST00000427769.1
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr18_+_56113488 | 1.45 |
ENST00000590797.1
|
RP11-1151B14.3
|
RP11-1151B14.3 |
| chr3_+_119499331 | 1.39 |
ENST00000393716.2
ENST00000466380.1 |
NR1I2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr1_+_22979474 | 1.35 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain |
| chr1_+_22979676 | 1.34 |
ENST00000432749.2
ENST00000314933.6 |
C1QB
|
complement component 1, q subcomponent, B chain |
| chr10_+_116853091 | 1.25 |
ENST00000526946.1
|
ATRNL1
|
attractin-like 1 |
| chr9_-_34710066 | 1.25 |
ENST00000378792.1
ENST00000259607.2 |
CCL21
|
chemokine (C-C motif) ligand 21 |
| chr2_+_115219171 | 1.24 |
ENST00000409163.1
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr1_-_62785054 | 1.24 |
ENST00000371153.4
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr22_+_23010756 | 1.18 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr1_-_62784935 | 1.17 |
ENST00000354381.3
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr12_+_101988774 | 1.17 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr5_-_54468974 | 1.17 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr11_+_124789146 | 1.12 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr12_+_103981044 | 1.11 |
ENST00000388887.2
|
STAB2
|
stabilin 2 |
| chr16_-_4838255 | 1.11 |
ENST00000591624.1
ENST00000396693.5 |
SEPT12
|
septin 12 |
| chr4_+_74275057 | 1.10 |
ENST00000511370.1
|
ALB
|
albumin |
| chr19_-_14168391 | 1.09 |
ENST00000589048.1
|
PALM3
|
paralemmin 3 |
| chr12_+_56075330 | 1.08 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr1_-_157567868 | 1.07 |
ENST00000271532.1
|
FCRL4
|
Fc receptor-like 4 |
| chr4_+_156775910 | 1.03 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr16_+_82068830 | 1.02 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr16_+_82068585 | 1.01 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr10_+_124030819 | 1.00 |
ENST00000260723.4
ENST00000368994.2 |
BTBD16
|
BTB (POZ) domain containing 16 |
| chr1_+_119911425 | 0.99 |
ENST00000361035.4
ENST00000325945.3 |
HAO2
|
hydroxyacid oxidase 2 (long chain) |
| chr4_+_74347400 | 0.96 |
ENST00000226355.3
|
AFM
|
afamin |
| chr5_-_115152651 | 0.94 |
ENST00000250535.4
|
CDO1
|
cysteine dioxygenase type 1 |
| chr12_+_101988627 | 0.94 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr1_+_110254850 | 0.93 |
ENST00000369812.5
ENST00000256593.3 ENST00000369813.1 |
GSTM5
|
glutathione S-transferase mu 5 |
| chr6_+_159291090 | 0.93 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr1_+_119911396 | 0.89 |
ENST00000457318.1
|
HAO2
|
hydroxyacid oxidase 2 (long chain) |
| chr14_+_94640671 | 0.88 |
ENST00000328839.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr16_+_56716336 | 0.87 |
ENST00000394485.4
ENST00000562939.1 |
MT1X
|
metallothionein 1X |
| chr16_+_82068873 | 0.87 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr1_+_85527987 | 0.86 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr12_-_47219733 | 0.86 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr1_+_196788887 | 0.84 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr7_+_100551239 | 0.84 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr11_-_5248294 | 0.81 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr5_-_42811986 | 0.81 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr7_+_302918 | 0.80 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr5_-_42812143 | 0.79 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr16_-_87970122 | 0.79 |
ENST00000309893.2
|
CA5A
|
carbonic anhydrase VA, mitochondrial |
| chr16_-_56701933 | 0.78 |
ENST00000568675.1
ENST00000569500.1 ENST00000444837.2 ENST00000379811.3 |
MT1G
|
metallothionein 1G |
| chr14_+_94640633 | 0.77 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr2_-_169887827 | 0.77 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr4_-_187517928 | 0.77 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chrX_-_52827141 | 0.76 |
ENST00000375511.3
|
SPANXN5
|
SPANX family, member N5 |
| chr5_-_101834617 | 0.75 |
ENST00000513675.1
ENST00000379807.3 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chrX_+_153813407 | 0.74 |
ENST00000443287.2
ENST00000333128.3 |
CTAG1A
|
cancer/testis antigen 1A |
| chr5_-_101834712 | 0.74 |
ENST00000506729.1
ENST00000389019.3 ENST00000379810.1 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr7_-_14026123 | 0.73 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr6_-_25930904 | 0.73 |
ENST00000377850.3
|
SLC17A2
|
solute carrier family 17, member 2 |
| chr11_-_914774 | 0.72 |
ENST00000528154.1
ENST00000525840.1 |
CHID1
|
chitinase domain containing 1 |
| chr22_-_27014043 | 0.72 |
ENST00000215939.2
|
CRYBB1
|
crystallin, beta B1 |
| chr17_-_14140166 | 0.71 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr6_+_160542870 | 0.71 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr5_+_176057663 | 0.71 |
ENST00000318682.6
|
EIF4E1B
|
eukaryotic translation initiation factor 4E family member 1B |
| chr2_-_26864228 | 0.70 |
ENST00000288861.4
|
CIB4
|
calcium and integrin binding family member 4 |
| chr10_-_48416849 | 0.70 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr17_-_34313685 | 0.70 |
ENST00000435911.2
ENST00000586216.1 ENST00000394509.4 |
CCL14
|
chemokine (C-C motif) ligand 14 |
| chr10_-_7513904 | 0.67 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr6_-_43478239 | 0.67 |
ENST00000372441.1
|
LRRC73
|
leucine rich repeat containing 73 |
| chr1_+_57320437 | 0.66 |
ENST00000361249.3
|
C8A
|
complement component 8, alpha polypeptide |
| chr5_-_176057518 | 0.65 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr12_+_24376201 | 0.65 |
ENST00000540733.1
ENST00000539583.1 |
RP11-778H2.1
|
RP11-778H2.1 |
| chr10_+_5005445 | 0.64 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr14_+_32798547 | 0.63 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr16_+_31539183 | 0.63 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chrX_-_5644225 | 0.63 |
ENST00000422914.1
|
RP11-733O18.1
|
RP11-733O18.1 |
| chr22_+_22556057 | 0.63 |
ENST00000390286.2
|
IGLV11-55
|
immunoglobulin lambda variable 11-55 (non-functional) |
| chr9_-_13432977 | 0.62 |
ENST00000605459.1
|
RP11-536O18.2
|
RP11-536O18.2 |
| chr16_+_31539197 | 0.62 |
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr5_-_176057365 | 0.62 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chrX_+_30260054 | 0.61 |
ENST00000378982.2
|
MAGEB4
|
melanoma antigen family B, 4 |
| chrX_+_69501943 | 0.60 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr12_-_22094336 | 0.60 |
ENST00000326684.4
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chrX_-_20134990 | 0.60 |
ENST00000379651.3
ENST00000443379.3 ENST00000379643.5 |
MAP7D2
|
MAP7 domain containing 2 |
| chr7_-_22234381 | 0.60 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chrX_+_140096761 | 0.59 |
ENST00000370530.1
|
SPANXB1
|
SPANX family, member B1 |
| chr22_+_44319648 | 0.59 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr6_+_131894284 | 0.59 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr19_-_40896081 | 0.59 |
ENST00000291823.2
|
HIPK4
|
homeodomain interacting protein kinase 4 |
| chr17_+_53343171 | 0.58 |
ENST00000430986.2
|
HLF
|
hepatic leukemia factor |
| chr12_-_127544894 | 0.58 |
ENST00000546062.1
ENST00000512624.2 ENST00000540244.1 |
RP11-575F12.1
|
RP11-575F12.1 |
| chr19_-_36297348 | 0.57 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr2_-_219858123 | 0.57 |
ENST00000453769.1
ENST00000295728.2 ENST00000392096.2 |
CRYBA2
|
crystallin, beta A2 |
| chr11_-_114271139 | 0.56 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr10_-_73497581 | 0.56 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr17_+_53343577 | 0.56 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr4_+_147560042 | 0.55 |
ENST00000281321.3
|
POU4F2
|
POU class 4 homeobox 2 |
| chr3_-_142720267 | 0.55 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr14_+_94577074 | 0.55 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr13_-_44453826 | 0.55 |
ENST00000444614.3
|
CCDC122
|
coiled-coil domain containing 122 |
| chr2_-_187713891 | 0.55 |
ENST00000295131.2
|
ZSWIM2
|
zinc finger, SWIM-type containing 2 |
| chr1_-_67600639 | 0.54 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr9_-_113100088 | 0.54 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr18_-_40695604 | 0.54 |
ENST00000590910.1
ENST00000326695.5 ENST00000589109.1 ENST00000282028.4 |
RIT2
|
Ras-like without CAAX 2 |
| chr16_+_76343743 | 0.53 |
ENST00000478060.1
|
CNTNAP4
|
contactin associated protein-like 4 |
| chr6_+_160542821 | 0.53 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr10_+_4448667 | 0.53 |
ENST00000434902.1
|
LINC00703
|
long intergenic non-protein coding RNA 703 |
| chr22_-_44708731 | 0.52 |
ENST00000381176.4
|
KIAA1644
|
KIAA1644 |
| chr7_-_14029515 | 0.52 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr17_+_30771279 | 0.52 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr1_-_109655355 | 0.52 |
ENST00000369945.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr7_-_14026063 | 0.51 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr2_-_166930131 | 0.51 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr1_-_109655377 | 0.50 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr10_-_13544945 | 0.50 |
ENST00000378605.3
ENST00000341083.3 |
BEND7
|
BEN domain containing 7 |
| chrX_-_139866723 | 0.50 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr9_-_138391692 | 0.50 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr11_+_58695174 | 0.49 |
ENST00000317391.4
|
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chr4_-_170924888 | 0.49 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr10_-_45496336 | 0.49 |
ENST00000298298.1
|
C10orf25
|
chromosome 10 open reading frame 25 |
| chr1_+_154966058 | 0.48 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr12_-_22094159 | 0.48 |
ENST00000538350.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_+_145549203 | 0.48 |
ENST00000355594.4
ENST00000544626.1 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr10_+_124320156 | 0.48 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr8_+_39972170 | 0.48 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr12_-_71533055 | 0.48 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr18_+_71983048 | 0.47 |
ENST00000579455.1
|
C18orf63
|
chromosome 18 open reading frame 63 |
| chr6_-_113953705 | 0.47 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr9_-_34376851 | 0.47 |
ENST00000297625.7
|
KIAA1161
|
KIAA1161 |
| chr2_+_171640291 | 0.47 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr10_+_122216316 | 0.46 |
ENST00000398250.1
ENST00000439221.1 ENST00000398248.1 |
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A |
| chr2_+_182850743 | 0.46 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_+_2005126 | 0.46 |
ENST00000495347.1
|
PRKCZ
|
protein kinase C, zeta |
| chr17_+_20483037 | 0.46 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
| chr7_-_122526799 | 0.46 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr1_-_169733789 | 0.46 |
ENST00000454271.1
ENST00000609271.1 |
RP1-117P20.3
SELE
|
RP1-117P20.3 selectin E |
| chr11_-_108338218 | 0.45 |
ENST00000525729.1
ENST00000393084.1 |
C11orf65
|
chromosome 11 open reading frame 65 |
| chr19_-_33555780 | 0.45 |
ENST00000254260.3
ENST00000400226.4 |
RHPN2
|
rhophilin, Rho GTPase binding protein 2 |
| chr8_-_121825088 | 0.45 |
ENST00000520717.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr3_+_153839149 | 0.45 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr11_-_10590238 | 0.45 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr8_+_20811140 | 0.45 |
ENST00000523035.1
|
RP11-369E15.4
|
RP11-369E15.4 |
| chr10_+_24498060 | 0.45 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr5_+_89854595 | 0.45 |
ENST00000405460.2
|
GPR98
|
G protein-coupled receptor 98 |
| chr6_+_132455526 | 0.44 |
ENST00000443303.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chrX_+_3189861 | 0.44 |
ENST00000457435.1
ENST00000420429.2 |
CXorf28
|
chromosome X open reading frame 28 |
| chr5_+_167181917 | 0.44 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr17_-_57604227 | 0.43 |
ENST00000584262.1
|
RP11-567L7.6
|
RP11-567L7.6 |
| chr22_+_21987005 | 0.43 |
ENST00000607942.1
ENST00000425975.1 ENST00000292779.3 |
CCDC116
|
coiled-coil domain containing 116 |
| chr7_+_1084206 | 0.42 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr1_+_197237352 | 0.42 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chrX_-_55187588 | 0.41 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr11_+_57308979 | 0.41 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr16_-_25122785 | 0.41 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr22_-_38380543 | 0.41 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr10_+_81370689 | 0.41 |
ENST00000372308.3
ENST00000398636.3 ENST00000428376.2 ENST00000372313.5 ENST00000419470.2 ENST00000429958.1 ENST00000439264.1 |
SFTPA1
|
surfactant protein A1 |
| chr15_-_20170354 | 0.41 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr2_-_175629135 | 0.41 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr22_+_25595817 | 0.41 |
ENST00000215855.2
ENST00000404334.1 |
CRYBB3
|
crystallin, beta B3 |
| chr21_-_47352477 | 0.41 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr11_+_74811578 | 0.41 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr5_-_13944652 | 0.41 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr1_-_177939348 | 0.40 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr6_+_46661575 | 0.40 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr1_-_177939041 | 0.40 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_87012753 | 0.40 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chrX_-_31090152 | 0.39 |
ENST00000359202.3
|
FTHL17
|
ferritin, heavy polypeptide-like 17 |
| chr7_-_22539771 | 0.39 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr17_+_9728828 | 0.39 |
ENST00000262441.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr14_-_47812321 | 0.39 |
ENST00000357362.3
ENST00000486952.2 ENST00000426342.1 |
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr3_+_50229037 | 0.39 |
ENST00000232461.3
ENST00000433068.1 |
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| chr12_-_91573132 | 0.39 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr10_-_90751038 | 0.39 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr19_+_48876300 | 0.38 |
ENST00000600863.1
ENST00000601610.1 ENST00000595322.1 |
SYNGR4
|
synaptogyrin 4 |
| chr17_+_27369918 | 0.38 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr11_+_60383204 | 0.38 |
ENST00000412599.1
ENST00000320202.4 |
LINC00301
|
long intergenic non-protein coding RNA 301 |
| chr1_-_203155868 | 0.37 |
ENST00000255409.3
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr9_+_34992846 | 0.37 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr16_-_25122735 | 0.37 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr14_-_23504087 | 0.36 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr15_+_90895471 | 0.36 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr4_+_154125565 | 0.36 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr11_-_110968081 | 0.36 |
ENST00000603154.1
|
RP11-89C3.4
|
RP11-89C3.4 |
| chr2_+_120189422 | 0.36 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr6_+_80816342 | 0.36 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr10_-_81320151 | 0.35 |
ENST00000372325.2
ENST00000372327.5 ENST00000417041.1 |
SFTPA2
|
surfactant protein A2 |
| chr11_+_22689648 | 0.35 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr7_+_117119187 | 0.35 |
ENST00000446805.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr5_+_110409012 | 0.35 |
ENST00000379706.4
|
TSLP
|
thymic stromal lymphopoietin |
| chr5_-_55412774 | 0.35 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr10_-_72542272 | 0.35 |
ENST00000545575.1
|
TBATA
|
thymus, brain and testes associated |
| chr16_+_4838393 | 0.35 |
ENST00000589721.1
|
SMIM22
|
small integral membrane protein 22 |
| chrX_-_154255143 | 0.34 |
ENST00000453950.1
ENST00000423959.1 |
F8
|
coagulation factor VIII, procoagulant component |
| chr15_+_85427903 | 0.34 |
ENST00000286749.3
ENST00000394573.1 ENST00000537703.1 |
SLC28A1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr8_-_27469383 | 0.34 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr16_+_4838412 | 0.33 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr8_-_110620284 | 0.33 |
ENST00000529690.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1903717 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.6 | 2.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.5 | 3.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 1.2 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.4 | 1.5 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.4 | 1.1 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.4 | 2.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 1.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.8 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.2 | 1.4 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.2 | 0.9 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 0.7 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.2 | 2.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 1.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 1.4 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.6 | GO:0035377 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.6 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.1 | 0.8 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 2.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.6 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.9 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.1 | 0.4 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.3 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.4 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.5 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.3 | GO:0003193 | pulmonary valve formation(GO:0003193) visceral motor neuron differentiation(GO:0021524) foramen ovale closure(GO:0035922) |
| 0.1 | 0.1 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.1 | 0.3 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.3 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 1.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 3.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.1 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.2 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 0.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.2 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.4 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 1.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 1.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.4 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 7.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.8 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.8 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.3 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.6 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.5 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.2 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.9 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0006090 | pyruvate metabolic process(GO:0006090) |
| 0.0 | 0.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.2 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.4 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.2 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.2 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.4 | GO:0021670 | lateral ventricle development(GO:0021670) outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 1.8 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 2.0 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 1.4 | GO:2000257 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.4 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.6 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.2 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 0.5 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 1.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.6 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 8.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 1.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 0.9 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 2.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 5.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.4 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.5 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.0 | GO:0034518 | nuclear cap binding complex(GO:0005846) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 4.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.6 | 3.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 1.9 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 2.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.3 | 1.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 0.8 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.3 | 2.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 1.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 1.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 0.7 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.1 | 0.6 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 1.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.6 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.5 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.1 | 0.4 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 3.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.7 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 1.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 8.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.3 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 1.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 2.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.2 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 1.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 0.4 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 1.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 1.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 6.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 3.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 7.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |