Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for MYBL1
Z-value: 0.10
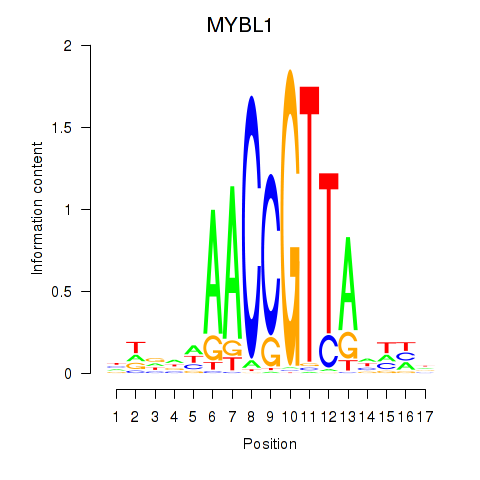
Transcription factors associated with MYBL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL1
|
ENSG00000185697.12 | MYB proto-oncogene like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL1 | hg19_v2_chr8_-_67525524_67525543 | 0.45 | 1.0e-02 | Click! |
Activity profile of MYBL1 motif
Sorted Z-values of MYBL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_16179884 | 2.73 |
ENST00000332432.8
|
MARCH11
|
membrane-associated ring finger (C3HC4) 11 |
| chr11_+_7110165 | 2.63 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chr11_+_45743858 | 2.61 |
ENST00000532307.1
|
CTD-2210P24.1
|
CTD-2210P24.1 |
| chrX_-_104465358 | 2.55 |
ENST00000372578.3
ENST00000372575.1 ENST00000413579.1 |
TEX13A
|
testis expressed 13A |
| chr19_+_44764031 | 2.54 |
ENST00000592581.1
ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233
|
zinc finger protein 233 |
| chr11_+_45743931 | 2.47 |
ENST00000530051.1
|
CTD-2210P24.1
|
CTD-2210P24.1 |
| chr2_-_132919528 | 2.32 |
ENST00000409867.1
|
ANKRD30BL
|
ankyrin repeat domain 30B-like |
| chr2_-_47382442 | 2.03 |
ENST00000445927.2
|
C2orf61
|
chromosome 2 open reading frame 61 |
| chr1_+_163291680 | 1.93 |
ENST00000450453.2
ENST00000524800.1 ENST00000442820.1 ENST00000367900.3 |
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr6_+_35748783 | 1.80 |
ENST00000373861.5
ENST00000542261.1 |
CLPSL1
|
colipase-like 1 |
| chr11_+_18433840 | 1.79 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr2_-_17699691 | 1.76 |
ENST00000399080.2
|
RAD51AP2
|
RAD51 associated protein 2 |
| chr17_+_58499066 | 1.75 |
ENST00000474834.1
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr8_+_25316707 | 1.71 |
ENST00000380665.3
|
CDCA2
|
cell division cycle associated 2 |
| chrX_-_140673133 | 1.65 |
ENST00000370519.3
|
SPANXA1
|
sperm protein associated with the nucleus, X-linked, family member A1 |
| chr1_+_92414928 | 1.64 |
ENST00000362005.3
ENST00000370389.2 ENST00000399546.2 ENST00000423434.1 ENST00000394530.3 ENST00000440509.1 |
BRDT
|
bromodomain, testis-specific |
| chr19_-_38397285 | 1.61 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chr8_+_25316489 | 1.60 |
ENST00000330560.3
|
CDCA2
|
cell division cycle associated 2 |
| chr1_+_173991633 | 1.58 |
ENST00000424181.1
|
RP11-160H22.3
|
RP11-160H22.3 |
| chr4_+_128651530 | 1.57 |
ENST00000281154.4
|
SLC25A31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr16_-_21289627 | 1.47 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chrX_+_147062844 | 1.32 |
ENST00000370467.3
|
FMR1NB
|
fragile X mental retardation 1 neighbor |
| chr22_+_19118321 | 1.31 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr7_-_18067478 | 1.30 |
ENST00000506618.2
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr10_+_124670121 | 1.25 |
ENST00000368894.1
|
FAM24A
|
family with sequence similarity 24, member A |
| chr16_+_67360856 | 1.25 |
ENST00000568804.2
|
LRRC36
|
leucine rich repeat containing 36 |
| chr8_-_91997427 | 1.23 |
ENST00000517562.2
|
RP11-122A3.2
|
chromosome 8 open reading frame 88 |
| chrX_+_105066524 | 1.16 |
ENST00000243300.9
ENST00000428173.2 |
NRK
|
Nik related kinase |
| chr19_-_55672037 | 1.15 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chrY_+_15815447 | 1.15 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chr4_-_177116772 | 1.14 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr6_-_27279949 | 1.12 |
ENST00000444565.1
ENST00000377451.2 |
POM121L2
|
POM121 transmembrane nucleoporin-like 2 |
| chr8_-_101157680 | 1.11 |
ENST00000428847.2
|
FBXO43
|
F-box protein 43 |
| chr6_+_74104471 | 1.10 |
ENST00000370336.4
|
DDX43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr8_+_50824233 | 1.07 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr16_+_89696692 | 1.05 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr14_+_23709555 | 1.03 |
ENST00000430154.2
|
C14orf164
|
chromosome 14 open reading frame 164 |
| chr19_-_9903666 | 0.99 |
ENST00000592587.1
|
ZNF846
|
zinc finger protein 846 |
| chr1_+_163291732 | 0.99 |
ENST00000271452.3
|
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr4_+_175839551 | 0.99 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr5_-_74162739 | 0.98 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr6_+_70576457 | 0.98 |
ENST00000322773.4
|
COL19A1
|
collagen, type XIX, alpha 1 |
| chr22_-_37403839 | 0.97 |
ENST00000402860.3
ENST00000381821.1 |
TEX33
|
testis expressed 33 |
| chr6_+_38683095 | 0.95 |
ENST00000449981.2
|
DNAH8
|
dynein, axonemal, heavy chain 8 |
| chr2_-_47382414 | 0.95 |
ENST00000294947.2
|
C2orf61
|
chromosome 2 open reading frame 61 |
| chr1_+_108918421 | 0.94 |
ENST00000444143.2
ENST00000294652.8 |
NBPF6
|
neuroblastoma breakpoint family, member 6 |
| chr12_+_104697504 | 0.94 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr2_-_74875432 | 0.91 |
ENST00000536235.1
ENST00000421985.1 |
M1AP
|
meiosis 1 associated protein |
| chr19_+_38826415 | 0.88 |
ENST00000410018.1
ENST00000409235.3 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr1_-_211848899 | 0.87 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr20_-_34117447 | 0.85 |
ENST00000246199.2
ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr7_+_63361201 | 0.84 |
ENST00000450544.1
|
RP11-340I6.8
|
RP11-340I6.8 |
| chr19_+_57640011 | 0.84 |
ENST00000598197.1
|
USP29
|
ubiquitin specific peptidase 29 |
| chr6_-_28554977 | 0.84 |
ENST00000452236.2
|
SCAND3
|
SCAN domain containing 3 |
| chr2_-_86564776 | 0.84 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr18_+_52385068 | 0.82 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chrX_+_37026432 | 0.82 |
ENST00000358047.3
|
FAM47C
|
family with sequence similarity 47, member C |
| chr9_+_104296243 | 0.82 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr22_-_37403858 | 0.81 |
ENST00000405091.2
|
TEX33
|
testis expressed 33 |
| chrX_-_70128580 | 0.81 |
ENST00000374333.2
|
TEX11
|
testis expressed 11 |
| chr5_-_74162605 | 0.80 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr20_-_56803695 | 0.79 |
ENST00000371167.3
ENST00000457363.1 |
ANKRD60
|
ankyrin repeat domain 60 |
| chr1_+_2066387 | 0.77 |
ENST00000497183.1
|
PRKCZ
|
protein kinase C, zeta |
| chr3_-_192445289 | 0.77 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr14_+_65007177 | 0.76 |
ENST00000247207.6
|
HSPA2
|
heat shock 70kDa protein 2 |
| chr3_+_50712672 | 0.75 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr11_+_93063137 | 0.74 |
ENST00000534747.1
|
CCDC67
|
coiled-coil domain containing 67 |
| chr17_-_56769382 | 0.74 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr19_+_58545434 | 0.73 |
ENST00000282326.1
ENST00000601162.1 |
ZSCAN1
|
zinc finger and SCAN domain containing 1 |
| chr10_+_91589261 | 0.73 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr17_-_46894576 | 0.73 |
ENST00000393382.3
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6 |
| chr2_-_86564696 | 0.73 |
ENST00000437769.1
|
REEP1
|
receptor accessory protein 1 |
| chr4_+_174818390 | 0.72 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chrX_+_71996972 | 0.71 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr2_-_64751227 | 0.70 |
ENST00000561559.1
|
RP11-568N6.1
|
RP11-568N6.1 |
| chr11_-_46638378 | 0.70 |
ENST00000529192.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr2_-_86564740 | 0.69 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr2_+_174219548 | 0.68 |
ENST00000347703.3
ENST00000392567.2 ENST00000306721.3 ENST00000410101.3 ENST00000410019.3 |
CDCA7
|
cell division cycle associated 7 |
| chr19_+_40503013 | 0.67 |
ENST00000595225.1
|
ZNF546
|
zinc finger protein 546 |
| chr1_+_85527987 | 0.66 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr9_-_35658007 | 0.65 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr19_-_13030071 | 0.65 |
ENST00000293695.7
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chr11_-_16419067 | 0.65 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr3_+_49236910 | 0.65 |
ENST00000452691.2
ENST00000366429.2 |
CCDC36
|
coiled-coil domain containing 36 |
| chr19_+_1495362 | 0.65 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr8_+_38244638 | 0.64 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr7_+_156433573 | 0.64 |
ENST00000311822.8
|
RNF32
|
ring finger protein 32 |
| chr6_+_13272904 | 0.63 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr13_-_49987885 | 0.63 |
ENST00000409082.1
|
CAB39L
|
calcium binding protein 39-like |
| chr16_+_4784273 | 0.63 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr11_+_64323156 | 0.63 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr4_+_2965307 | 0.63 |
ENST00000398051.4
ENST00000503518.2 ENST00000398052.4 ENST00000345167.6 ENST00000504933.1 ENST00000442472.2 |
GRK4
|
G protein-coupled receptor kinase 4 |
| chrY_-_21906594 | 0.62 |
ENST00000447300.1
|
KDM5D
|
lysine (K)-specific demethylase 5D |
| chr15_+_66797627 | 0.62 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chrX_+_105066554 | 0.61 |
ENST00000536164.1
|
NRK
|
Nik related kinase |
| chr18_-_71815051 | 0.61 |
ENST00000582526.1
ENST00000419743.2 |
FBXO15
|
F-box protein 15 |
| chr1_+_179335101 | 0.60 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
| chr19_+_36239576 | 0.60 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr19_+_37096194 | 0.60 |
ENST00000460670.1
ENST00000292928.2 ENST00000439428.1 |
ZNF382
|
zinc finger protein 382 |
| chr3_+_173116225 | 0.60 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr9_+_104296163 | 0.59 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chrY_-_21239221 | 0.59 |
ENST00000447937.1
ENST00000331787.2 |
TTTY14
|
testis-specific transcript, Y-linked 14 (non-protein coding) |
| chr20_-_52687030 | 0.59 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr1_-_205326161 | 0.58 |
ENST00000367156.3
ENST00000606887.1 ENST00000607173.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr1_-_247921982 | 0.58 |
ENST00000408896.2
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1 |
| chr1_-_68915610 | 0.57 |
ENST00000262340.5
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa |
| chr18_-_48346130 | 0.57 |
ENST00000592966.1
|
MRO
|
maestro |
| chr5_-_95018660 | 0.57 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr19_-_58485895 | 0.56 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr6_-_110797642 | 0.56 |
ENST00000434949.1
|
SLC22A16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr22_+_32455111 | 0.56 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr3_-_49893907 | 0.56 |
ENST00000482582.1
|
TRAIP
|
TRAF interacting protein |
| chr18_-_48346298 | 0.56 |
ENST00000398439.3
|
MRO
|
maestro |
| chr2_+_95963052 | 0.55 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr18_-_44497308 | 0.54 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr18_-_71814999 | 0.54 |
ENST00000269500.5
|
FBXO15
|
F-box protein 15 |
| chr1_+_245027971 | 0.53 |
ENST00000610145.1
|
RP11-11N7.4
|
RP11-11N7.4 |
| chr3_+_12598563 | 0.53 |
ENST00000411987.1
ENST00000448482.1 |
MKRN2
|
makorin ring finger protein 2 |
| chr8_+_105235572 | 0.53 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr11_+_64323098 | 0.52 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chrX_+_49969405 | 0.52 |
ENST00000376042.1
|
CCNB3
|
cyclin B3 |
| chr1_-_68962805 | 0.51 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr19_+_38826477 | 0.50 |
ENST00000409410.2
ENST00000215069.4 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr1_-_190443931 | 0.50 |
ENST00000445957.2
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr15_+_66797455 | 0.50 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr13_-_32889477 | 0.50 |
ENST00000533490.2
|
ZAR1L
|
zygote arrest 1-like |
| chr6_+_26240561 | 0.50 |
ENST00000377745.2
|
HIST1H4F
|
histone cluster 1, H4f |
| chr18_-_48346415 | 0.50 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr20_-_52687059 | 0.50 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr3_-_148804275 | 0.49 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr19_+_52839490 | 0.49 |
ENST00000321287.8
|
ZNF610
|
zinc finger protein 610 |
| chr19_-_37096139 | 0.48 |
ENST00000585983.1
ENST00000585960.1 ENST00000586115.1 |
ZNF529
|
zinc finger protein 529 |
| chr19_-_44258733 | 0.48 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr3_+_63428982 | 0.48 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr19_-_38183228 | 0.48 |
ENST00000587199.1
|
ZFP30
|
ZFP30 zinc finger protein |
| chr12_+_49717019 | 0.47 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr19_-_19144243 | 0.46 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr19_-_55919087 | 0.46 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr9_+_104296122 | 0.46 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr17_+_47865917 | 0.46 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr4_-_4544061 | 0.45 |
ENST00000507908.1
|
STX18
|
syntaxin 18 |
| chr1_-_63782888 | 0.45 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr16_-_29934558 | 0.44 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr20_+_5931497 | 0.44 |
ENST00000378886.2
ENST00000265187.4 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr1_-_247876105 | 0.44 |
ENST00000302084.2
|
OR6F1
|
olfactory receptor, family 6, subfamily F, member 1 |
| chr4_+_184826418 | 0.44 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr12_+_69753448 | 0.43 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr6_-_123958111 | 0.43 |
ENST00000542443.1
|
TRDN
|
triadin |
| chr14_+_58894404 | 0.43 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr1_+_44889697 | 0.43 |
ENST00000443020.2
|
RNF220
|
ring finger protein 220 |
| chr11_-_27723158 | 0.43 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr11_+_125757556 | 0.43 |
ENST00000526028.1
|
HYLS1
|
hydrolethalus syndrome 1 |
| chr19_-_6199521 | 0.43 |
ENST00000592883.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr7_+_100547156 | 0.43 |
ENST00000379458.4
|
MUC3A
|
Protein LOC100131514 |
| chr11_+_33037652 | 0.43 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr2_-_208994548 | 0.42 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr16_-_25122735 | 0.42 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr1_-_39395165 | 0.42 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr1_+_92414952 | 0.41 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr4_-_120988229 | 0.41 |
ENST00000296509.6
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr5_-_137514617 | 0.41 |
ENST00000254900.5
|
BRD8
|
bromodomain containing 8 |
| chr6_-_29324054 | 0.41 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr15_-_43212836 | 0.41 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr5_+_35617940 | 0.41 |
ENST00000282469.6
ENST00000509059.1 ENST00000356031.3 ENST00000510777.1 |
SPEF2
|
sperm flagellar 2 |
| chr8_+_95565947 | 0.40 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr1_+_155829286 | 0.40 |
ENST00000368324.4
|
SYT11
|
synaptotagmin XI |
| chr3_+_100120441 | 0.40 |
ENST00000489752.1
|
LNP1
|
leukemia NUP98 fusion partner 1 |
| chr5_+_94982435 | 0.39 |
ENST00000511684.1
ENST00000380005.4 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr18_-_70211691 | 0.39 |
ENST00000269503.4
|
CBLN2
|
cerebellin 2 precursor |
| chr19_+_40502938 | 0.39 |
ENST00000599504.1
ENST00000596894.1 ENST00000601138.1 ENST00000600094.1 ENST00000347077.4 |
ZNF546
|
zinc finger protein 546 |
| chr1_+_97187318 | 0.39 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr9_+_34458851 | 0.39 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr13_+_32889605 | 0.38 |
ENST00000380152.3
ENST00000544455.1 ENST00000530893.2 |
BRCA2
|
breast cancer 2, early onset |
| chr15_-_43212996 | 0.38 |
ENST00000567840.1
|
TTBK2
|
tau tubulin kinase 2 |
| chr1_+_172502244 | 0.38 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr10_-_28623368 | 0.38 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr14_+_58894103 | 0.37 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr20_-_23969416 | 0.36 |
ENST00000335694.4
|
GGTLC1
|
gamma-glutamyltransferase light chain 1 |
| chr7_-_34978980 | 0.36 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr3_-_190167571 | 0.36 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr2_+_122513109 | 0.36 |
ENST00000389682.3
ENST00000536142.1 |
TSN
|
translin |
| chr1_-_159869912 | 0.35 |
ENST00000368099.4
|
CCDC19
|
coiled-coil domain containing 19 |
| chr10_-_82116497 | 0.35 |
ENST00000372204.3
|
DYDC1
|
DPY30 domain containing 1 |
| chr22_+_43011247 | 0.35 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr4_-_76649546 | 0.34 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr7_+_12726474 | 0.34 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr8_-_82598067 | 0.34 |
ENST00000523942.1
ENST00000522997.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr3_-_10052869 | 0.34 |
ENST00000454232.1
|
AC022007.5
|
AC022007.5 |
| chr1_-_205325994 | 0.34 |
ENST00000491471.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr1_+_45805728 | 0.34 |
ENST00000539779.1
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr1_-_197115818 | 0.34 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr6_+_134274354 | 0.34 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr17_-_30668887 | 0.33 |
ENST00000581747.1
ENST00000583334.1 ENST00000580558.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr19_-_38183201 | 0.33 |
ENST00000590008.1
ENST00000358582.4 |
ZNF781
|
zinc finger protein 781 |
| chr1_+_94798754 | 0.33 |
ENST00000418242.1
|
RP11-148B18.3
|
RP11-148B18.3 |
| chr3_-_182703688 | 0.32 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr13_-_25746416 | 0.32 |
ENST00000515384.1
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr19_-_6110555 | 0.32 |
ENST00000593241.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr14_+_58894141 | 0.32 |
ENST00000423743.3
|
KIAA0586
|
KIAA0586 |
| chr18_-_3880051 | 0.32 |
ENST00000584874.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.7 | 2.1 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.6 | 1.8 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.4 | 1.8 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.4 | 1.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.3 | 1.7 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.3 | 1.3 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 0.6 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.2 | 1.9 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 | 0.7 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 0.5 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.6 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 0.4 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.4 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 0.9 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.9 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.5 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.6 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.1 | 1.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.6 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.3 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 2.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.3 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 1.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 0.7 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 1.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.6 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.1 | 1.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.5 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 1.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.6 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:1903947 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.4 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.4 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 2.0 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 1.3 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 1.4 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.5 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.8 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 2.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 2.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.3 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.4 | 2.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.3 | 1.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 2.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 1.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.5 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.4 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 2.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.8 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 3.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 3.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.3 | 2.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 0.6 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 1.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.8 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 1.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.5 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 1.1 | GO:0034235 | GPI anchor binding(GO:0034235) metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.3 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.1 | 0.6 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 1.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.6 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.2 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 1.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 3.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 2.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |