Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for NKX1-1
Z-value: 0.21
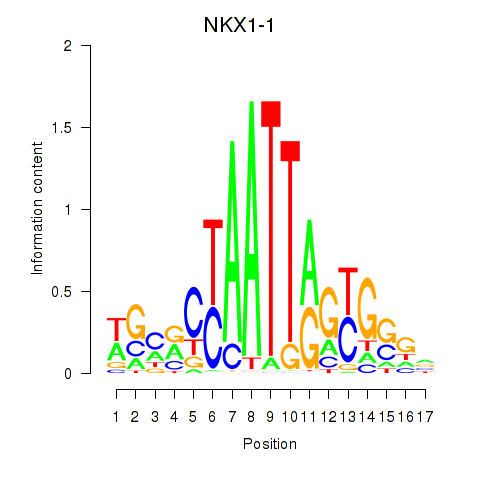
Transcription factors associated with NKX1-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX1-1
|
ENSG00000235608.1 | NK1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX1-1 | hg19_v2_chr4_-_1400119_1400119 | -0.84 | 1.3e-09 | Click! |
Activity profile of NKX1-1 motif
Sorted Z-values of NKX1-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_16500599 | 3.11 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr12_+_16500571 | 3.09 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr12_+_16500037 | 2.98 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr3_+_186560476 | 1.32 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr3_+_186560462 | 1.31 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr10_+_5238793 | 0.89 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr14_+_22670455 | 0.83 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr9_+_34646651 | 0.81 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr7_+_26331541 | 0.80 |
ENST00000416246.1
ENST00000338523.4 ENST00000412416.1 |
SNX10
|
sorting nexin 10 |
| chr17_-_27418537 | 0.75 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr12_+_64798826 | 0.69 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr7_+_26331678 | 0.69 |
ENST00000446848.2
|
SNX10
|
sorting nexin 10 |
| chr9_+_34646624 | 0.67 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr10_+_120789223 | 0.66 |
ENST00000425699.1
|
NANOS1
|
nanos homolog 1 (Drosophila) |
| chr1_+_161691353 | 0.65 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr7_+_50348268 | 0.64 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr4_-_89442940 | 0.64 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chrX_+_19362011 | 0.60 |
ENST00000379806.5
ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr13_-_36050819 | 0.59 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr2_-_47143160 | 0.58 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr1_-_21620877 | 0.55 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr2_-_47142884 | 0.52 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr7_+_2559399 | 0.50 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr22_+_22730353 | 0.50 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr6_-_167040693 | 0.48 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr6_-_82957433 | 0.47 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr19_-_14228541 | 0.47 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr22_-_39151463 | 0.46 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr22_+_41601209 | 0.46 |
ENST00000216237.5
|
L3MBTL2
|
l(3)mbt-like 2 (Drosophila) |
| chr2_+_8822113 | 0.46 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr5_+_66300464 | 0.46 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr10_+_99205894 | 0.45 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr3_+_158519654 | 0.44 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr10_-_99205607 | 0.44 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr17_+_57233087 | 0.43 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr10_+_99205959 | 0.41 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr1_-_221915418 | 0.41 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr14_-_24711764 | 0.39 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr11_-_124981475 | 0.38 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr9_+_116225999 | 0.37 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr7_-_6048650 | 0.37 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr11_-_47198380 | 0.37 |
ENST00000419701.2
ENST00000526342.1 ENST00000528444.1 ENST00000530596.1 ENST00000525398.1 ENST00000319543.6 ENST00000426335.2 ENST00000527927.1 ENST00000525314.1 |
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr1_+_155278539 | 0.37 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chrX_+_55026763 | 0.37 |
ENST00000374987.3
|
APEX2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr10_+_1095416 | 0.37 |
ENST00000358220.1
|
WDR37
|
WD repeat domain 37 |
| chr16_-_11036300 | 0.37 |
ENST00000331808.4
|
DEXI
|
Dexi homolog (mouse) |
| chr1_+_62439037 | 0.36 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr5_+_66300446 | 0.36 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_-_24711806 | 0.35 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr1_+_155278625 | 0.35 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr21_-_37914898 | 0.35 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr3_-_105587879 | 0.35 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr17_+_45286387 | 0.34 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr11_-_62521614 | 0.34 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr17_+_57232690 | 0.33 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr11_-_65625678 | 0.32 |
ENST00000308162.5
|
CFL1
|
cofilin 1 (non-muscle) |
| chr14_-_24711470 | 0.32 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr15_-_78526942 | 0.32 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr6_-_167040731 | 0.31 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr14_-_24711865 | 0.31 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr18_+_55888767 | 0.30 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr17_+_45286706 | 0.30 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr5_-_115890554 | 0.29 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_-_1094819 | 0.28 |
ENST00000429642.1
|
IDI1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr12_+_28410128 | 0.28 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr4_-_54232144 | 0.28 |
ENST00000388940.4
ENST00000503450.1 ENST00000401642.3 |
SCFD2
|
sec1 family domain containing 2 |
| chr17_-_8055747 | 0.28 |
ENST00000317276.4
ENST00000581703.1 |
PER1
|
period circadian clock 1 |
| chr17_-_45266542 | 0.27 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr2_+_105050794 | 0.26 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr15_+_96904487 | 0.26 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr10_-_1095050 | 0.25 |
ENST00000381344.3
|
IDI1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr12_+_48499252 | 0.25 |
ENST00000549003.1
ENST00000550924.1 |
PFKM
|
phosphofructokinase, muscle |
| chr19_+_50270219 | 0.25 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr7_+_99647411 | 0.25 |
ENST00000438937.1
|
ZSCAN21
|
zinc finger and SCAN domain containing 21 |
| chrX_-_153363125 | 0.25 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr7_-_6048702 | 0.24 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr18_+_63273310 | 0.23 |
ENST00000579862.1
|
RP11-775G23.1
|
RP11-775G23.1 |
| chr11_-_62494821 | 0.23 |
ENST00000301785.5
|
HNRNPUL2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr17_-_57232596 | 0.23 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr14_+_32798462 | 0.22 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr7_-_33102338 | 0.22 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr7_-_33102399 | 0.21 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr11_-_59383617 | 0.21 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr12_-_102513843 | 0.21 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr17_-_10372875 | 0.20 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr7_+_6048856 | 0.20 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr12_+_2921788 | 0.19 |
ENST00000228799.2
ENST00000419778.2 ENST00000542548.1 |
ITFG2
|
integrin alpha FG-GAP repeat containing 2 |
| chr3_-_105588231 | 0.19 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr11_-_111741994 | 0.19 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr15_-_75199213 | 0.19 |
ENST00000562698.1
|
FAM219B
|
family with sequence similarity 219, member B |
| chr4_+_128554081 | 0.18 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr8_+_94929077 | 0.18 |
ENST00000297598.4
ENST00000520614.1 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr8_+_26371763 | 0.18 |
ENST00000521913.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr19_-_6424783 | 0.18 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr3_+_186288454 | 0.18 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr1_-_159915386 | 0.17 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr2_-_97405775 | 0.17 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr8_+_94929168 | 0.16 |
ENST00000518107.1
ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_-_57232525 | 0.16 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_+_119205222 | 0.15 |
ENST00000311413.4
|
RNF26
|
ring finger protein 26 |
| chr15_-_64386120 | 0.15 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr4_-_103682145 | 0.15 |
ENST00000226578.4
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr3_+_57261743 | 0.15 |
ENST00000288266.3
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr11_+_72983246 | 0.14 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr7_+_99647389 | 0.14 |
ENST00000543588.1
ENST00000292450.4 ENST00000456748.2 |
ZSCAN21
|
zinc finger and SCAN domain containing 21 |
| chr8_+_94929110 | 0.14 |
ENST00000520728.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_-_108507631 | 0.14 |
ENST00000527011.1
ENST00000370056.4 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr5_+_150226085 | 0.13 |
ENST00000522154.1
|
IRGM
|
immunity-related GTPase family, M |
| chr2_+_187371440 | 0.13 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr17_-_44657017 | 0.13 |
ENST00000573185.1
ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A
|
ADP-ribosylation factor-like 17A |
| chr11_-_118023594 | 0.12 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr12_+_6961279 | 0.12 |
ENST00000229268.8
ENST00000389231.5 ENST00000542087.1 |
USP5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr17_-_61777459 | 0.12 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr17_-_44439084 | 0.12 |
ENST00000575960.1
ENST00000575698.1 ENST00000571246.1 ENST00000434041.2 ENST00000570618.1 ENST00000450673.3 |
ARL17B
|
ADP-ribosylation factor-like 17B |
| chrX_-_47509994 | 0.11 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chr9_+_124329336 | 0.11 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr2_-_167232484 | 0.11 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr1_-_231004220 | 0.11 |
ENST00000366663.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr3_+_23851928 | 0.11 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr11_-_71823796 | 0.10 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr8_+_98656693 | 0.10 |
ENST00000519934.1
|
MTDH
|
metadherin |
| chr15_-_64385981 | 0.10 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr19_+_45504688 | 0.10 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr1_+_222910625 | 0.10 |
ENST00000360827.2
|
FAM177B
|
family with sequence similarity 177, member B |
| chr11_-_4629388 | 0.10 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr4_-_19458597 | 0.09 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr17_-_8113886 | 0.09 |
ENST00000577833.1
ENST00000534871.1 ENST00000583915.1 ENST00000316199.6 ENST00000581511.1 ENST00000585124.1 |
AURKB
|
aurora kinase B |
| chr5_-_159546396 | 0.09 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr11_-_4629367 | 0.08 |
ENST00000533021.1
|
TRIM68
|
tripartite motif containing 68 |
| chr11_-_71823715 | 0.08 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr6_-_85473073 | 0.07 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr19_+_36249057 | 0.06 |
ENST00000301165.5
ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr1_+_46972668 | 0.06 |
ENST00000371956.4
ENST00000360032.3 |
DMBX1
|
diencephalon/mesencephalon homeobox 1 |
| chr13_+_36050881 | 0.05 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr7_-_27239703 | 0.05 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr19_-_8070474 | 0.05 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chr16_-_66864806 | 0.05 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr11_-_105892937 | 0.05 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr10_-_56560939 | 0.04 |
ENST00000373955.1
|
PCDH15
|
protocadherin-related 15 |
| chr8_+_98656336 | 0.04 |
ENST00000336273.3
|
MTDH
|
metadherin |
| chr9_+_139780942 | 0.03 |
ENST00000247668.2
ENST00000359662.3 |
TRAF2
|
TNF receptor-associated factor 2 |
| chrX_-_47509887 | 0.03 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chrX_+_72783026 | 0.03 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr7_-_99716914 | 0.03 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_+_174843548 | 0.02 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr8_+_94929273 | 0.02 |
ENST00000518573.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_-_40337470 | 0.00 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX1-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.9 | 2.6 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.5 | 1.5 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.2 | 1.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.7 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.5 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.1 | 0.9 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.6 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.3 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 1.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.5 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.6 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.7 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.9 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.6 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 1.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 9.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 0.9 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.2 | 2.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 0.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.7 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.5 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 0.6 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.5 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.4 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.6 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 0.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.4 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 2.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |