Project
Illumina Body Map 2, young vs old
Navigation
Downloads
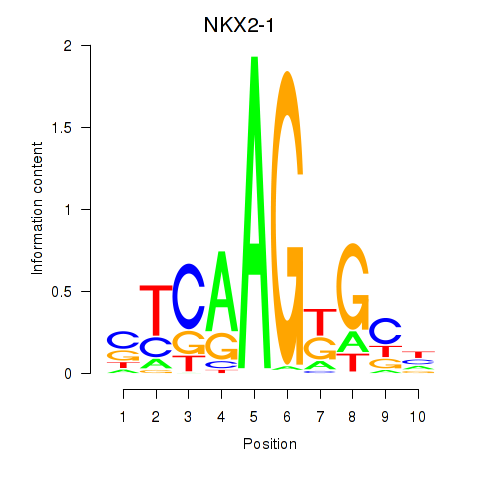
Results for NKX2-1
Z-value: 0.73
Transcription factors associated with NKX2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-1
|
ENSG00000136352.13 | NK2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-1 | hg19_v2_chr14_-_36990061_36990154 | 0.56 | 9.4e-04 | Click! |
Activity profile of NKX2-1 motif
Sorted Z-values of NKX2-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_101962128 | 5.97 |
ENST00000550514.1
|
MYBPC1
|
myosin binding protein C, slow type |
| chr3_-_42744312 | 3.44 |
ENST00000416756.1
ENST00000441594.1 |
HHATL
|
hedgehog acyltransferase-like |
| chr20_+_30407151 | 3.25 |
ENST00000375985.4
|
MYLK2
|
myosin light chain kinase 2 |
| chr20_+_30407105 | 3.17 |
ENST00000375994.2
|
MYLK2
|
myosin light chain kinase 2 |
| chr17_-_10452929 | 3.11 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr19_+_38924316 | 3.10 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chr9_-_97356075 | 2.80 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr2_+_168043793 | 2.43 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr8_+_22019168 | 2.38 |
ENST00000318561.3
ENST00000521315.1 ENST00000437090.2 ENST00000520605.1 ENST00000522109.1 ENST00000524255.1 ENST00000523296.1 ENST00000518615.1 |
SFTPC
|
surfactant protein C |
| chr3_-_42744270 | 1.85 |
ENST00000457462.1
|
HHATL
|
hedgehog acyltransferase-like |
| chr6_-_113953705 | 1.84 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr3_-_42744130 | 1.74 |
ENST00000417472.1
ENST00000442469.1 |
HHATL
|
hedgehog acyltransferase-like |
| chr15_-_42448788 | 1.74 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr10_-_81320151 | 1.67 |
ENST00000372325.2
ENST00000372327.5 ENST00000417041.1 |
SFTPA2
|
surfactant protein A2 |
| chr6_-_117747015 | 1.61 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr17_-_1508379 | 1.53 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr3_+_121311966 | 1.45 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr14_+_81421710 | 1.43 |
ENST00000342443.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chr16_+_28303804 | 1.39 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr14_+_81421355 | 1.33 |
ENST00000541158.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr14_+_81421921 | 1.28 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chrX_+_22056165 | 1.20 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr14_+_81421861 | 1.19 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr5_-_58335281 | 1.11 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_58571935 | 1.11 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr14_+_94393605 | 1.08 |
ENST00000556222.1
ENST00000554404.1 ENST00000557000.2 |
FAM181A
|
family with sequence similarity 181, member A |
| chr2_+_220299547 | 1.06 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr2_-_85895295 | 1.06 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr17_-_42466864 | 0.98 |
ENST00000353281.4
ENST00000262407.5 |
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr6_+_43968306 | 0.97 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr17_+_59529743 | 0.94 |
ENST00000589003.1
ENST00000393853.4 |
TBX4
|
T-box 4 |
| chr14_+_22362613 | 0.92 |
ENST00000390438.2
|
TRAV8-4
|
T cell receptor alpha variable 8-4 |
| chr11_+_60223225 | 0.90 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr7_-_24957699 | 0.85 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr6_-_160679905 | 0.84 |
ENST00000366953.3
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr10_+_63808970 | 0.83 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr3_+_155755482 | 0.82 |
ENST00000472028.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr11_+_35160709 | 0.79 |
ENST00000415148.2
ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44
|
CD44 molecule (Indian blood group) |
| chrX_-_108976410 | 0.78 |
ENST00000504980.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr3_-_139195350 | 0.78 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr13_-_41240717 | 0.76 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr17_-_49198216 | 0.75 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr6_-_134373732 | 0.73 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chrX_-_108976449 | 0.73 |
ENST00000469857.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr15_+_74911430 | 0.73 |
ENST00000562670.1
ENST00000564096.1 |
CLK3
|
CDC-like kinase 3 |
| chr6_-_41673552 | 0.72 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr1_-_17338386 | 0.71 |
ENST00000341676.5
ENST00000452699.1 |
ATP13A2
|
ATPase type 13A2 |
| chr19_+_55014085 | 0.70 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr19_-_10679644 | 0.70 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr16_+_640055 | 0.69 |
ENST00000568586.1
ENST00000538492.1 ENST00000248139.3 |
RAB40C
|
RAB40C, member RAS oncogene family |
| chr9_+_109685630 | 0.68 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr20_+_825275 | 0.67 |
ENST00000541082.1
|
FAM110A
|
family with sequence similarity 110, member A |
| chr12_-_54694758 | 0.65 |
ENST00000553070.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr17_-_76778339 | 0.65 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr6_-_87804815 | 0.64 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr14_-_106068065 | 0.63 |
ENST00000390541.2
|
IGHE
|
immunoglobulin heavy constant epsilon |
| chr22_-_30968813 | 0.63 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr22_-_30968839 | 0.61 |
ENST00000445645.1
ENST00000416358.1 ENST00000423371.1 ENST00000411821.1 ENST00000448604.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr1_-_153538011 | 0.60 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr6_-_26056695 | 0.59 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr11_-_105948040 | 0.56 |
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr11_+_60223312 | 0.56 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr6_+_31674639 | 0.56 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr17_+_40610862 | 0.56 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr12_-_719573 | 0.55 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr15_-_94614049 | 0.54 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr8_-_16859690 | 0.54 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chrX_+_23928500 | 0.54 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr9_+_33290491 | 0.53 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr1_+_66999268 | 0.53 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr1_-_153538292 | 0.52 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr3_-_56950407 | 0.51 |
ENST00000496106.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr8_-_114389353 | 0.48 |
ENST00000343508.3
|
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr1_-_175712665 | 0.47 |
ENST00000263525.2
|
TNR
|
tenascin R |
| chr8_+_145203548 | 0.47 |
ENST00000534366.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr1_-_228604328 | 0.46 |
ENST00000355586.4
ENST00000366698.2 ENST00000520264.1 ENST00000479800.1 ENST00000295033.3 |
TRIM17
|
tripartite motif containing 17 |
| chr3_-_11685345 | 0.45 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr7_-_142583506 | 0.44 |
ENST00000359396.3
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr14_-_74892805 | 0.44 |
ENST00000331628.3
ENST00000554953.1 |
SYNDIG1L
|
synapse differentiation inducing 1-like |
| chr21_+_45773515 | 0.44 |
ENST00000397932.2
ENST00000300481.9 |
TRPM2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr9_+_129986734 | 0.44 |
ENST00000444677.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr8_-_144679602 | 0.43 |
ENST00000526710.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr19_-_39390212 | 0.43 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr21_-_36421401 | 0.43 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr19_-_51336443 | 0.42 |
ENST00000598673.1
|
KLK15
|
kallikrein-related peptidase 15 |
| chr15_-_102463298 | 0.42 |
ENST00000326183.3
|
OR4F4
|
olfactory receptor, family 4, subfamily F, member 4 |
| chr15_+_76030311 | 0.42 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr11_-_105948129 | 0.41 |
ENST00000526793.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr5_+_131705438 | 0.40 |
ENST00000245407.3
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr10_+_111967345 | 0.39 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr1_-_175712829 | 0.39 |
ENST00000367674.2
|
TNR
|
tenascin R |
| chr3_+_19189946 | 0.39 |
ENST00000328405.2
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr17_+_61554829 | 0.38 |
ENST00000582627.1
|
ACE
|
angiotensin I converting enzyme |
| chr11_-_15643937 | 0.38 |
ENST00000533082.1
|
RP11-531H8.2
|
RP11-531H8.2 |
| chr9_+_133986782 | 0.37 |
ENST00000372301.2
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr6_-_71012773 | 0.37 |
ENST00000370496.3
ENST00000357250.6 |
COL9A1
|
collagen, type IX, alpha 1 |
| chr5_-_134783038 | 0.36 |
ENST00000503143.2
|
C5orf20
|
chromosome 5 open reading frame 20 |
| chr8_+_118147498 | 0.35 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr8_+_70404996 | 0.34 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr14_+_67708137 | 0.34 |
ENST00000556345.1
ENST00000555925.1 ENST00000557783.1 |
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr15_-_33360085 | 0.32 |
ENST00000334528.9
|
FMN1
|
formin 1 |
| chr18_+_13382553 | 0.32 |
ENST00000586222.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr19_-_39390350 | 0.31 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr6_-_42858534 | 0.31 |
ENST00000408925.2
|
C6orf226
|
chromosome 6 open reading frame 226 |
| chr20_-_35724388 | 0.30 |
ENST00000344359.3
ENST00000373664.3 |
RBL1
|
retinoblastoma-like 1 (p107) |
| chr16_+_640201 | 0.30 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr16_-_18937072 | 0.30 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr19_+_48972459 | 0.29 |
ENST00000427476.1
|
CYTH2
|
cytohesin 2 |
| chr17_-_34195889 | 0.29 |
ENST00000311880.2
|
C17orf66
|
chromosome 17 open reading frame 66 |
| chr10_-_103454876 | 0.29 |
ENST00000331272.7
|
FBXW4
|
F-box and WD repeat domain containing 4 |
| chr2_+_220379052 | 0.28 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr19_+_50148087 | 0.28 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr19_+_48972265 | 0.27 |
ENST00000452733.2
|
CYTH2
|
cytohesin 2 |
| chr17_-_37123646 | 0.27 |
ENST00000378079.2
|
FBXO47
|
F-box protein 47 |
| chr7_-_27224795 | 0.27 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chrX_+_136648297 | 0.27 |
ENST00000287538.5
|
ZIC3
|
Zic family member 3 |
| chr7_-_142583478 | 0.26 |
ENST00000436401.1
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr10_+_82300575 | 0.26 |
ENST00000313455.4
|
SH2D4B
|
SH2 domain containing 4B |
| chr12_+_56401268 | 0.26 |
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr1_-_247876105 | 0.26 |
ENST00000302084.2
|
OR6F1
|
olfactory receptor, family 6, subfamily F, member 1 |
| chr4_+_55524085 | 0.25 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr10_+_102222798 | 0.25 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr16_+_2022036 | 0.25 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr19_-_38720354 | 0.25 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr1_-_156269428 | 0.24 |
ENST00000339922.3
|
VHLL
|
von Hippel-Lindau tumor suppressor-like |
| chr4_-_122872029 | 0.24 |
ENST00000502968.1
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr12_+_13349711 | 0.24 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr6_+_42883727 | 0.24 |
ENST00000304672.1
ENST00000441198.1 ENST00000446507.1 |
PTCRA
|
pre T-cell antigen receptor alpha |
| chr16_-_67493110 | 0.23 |
ENST00000602876.1
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr17_-_34195862 | 0.23 |
ENST00000592980.1
ENST00000587626.1 |
C17orf66
|
chromosome 17 open reading frame 66 |
| chr1_+_204485571 | 0.23 |
ENST00000454264.2
ENST00000367183.3 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr20_-_56285595 | 0.23 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr11_+_105948216 | 0.23 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr2_-_105882585 | 0.23 |
ENST00000595531.1
|
AC012360.2
|
LOC644617 protein; Uncharacterized protein |
| chr3_+_184056614 | 0.22 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr12_+_25205628 | 0.22 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr7_-_27224842 | 0.22 |
ENST00000517402.1
|
HOXA11
|
homeobox A11 |
| chr1_+_206643806 | 0.22 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr12_+_121837905 | 0.22 |
ENST00000392465.3
ENST00000554606.1 ENST00000392464.2 ENST00000555076.1 |
RNF34
|
ring finger protein 34, E3 ubiquitin protein ligase |
| chr14_+_67707826 | 0.22 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr5_-_172662303 | 0.22 |
ENST00000517440.1
ENST00000329198.4 |
NKX2-5
|
NK2 homeobox 5 |
| chr6_-_55740352 | 0.22 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr3_+_4535025 | 0.22 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr12_-_21810726 | 0.21 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr22_+_50986462 | 0.21 |
ENST00000395676.2
|
KLHDC7B
|
kelch domain containing 7B |
| chr19_+_1942453 | 0.21 |
ENST00000591752.1
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr17_+_59489112 | 0.21 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr10_-_98031155 | 0.21 |
ENST00000495266.1
|
BLNK
|
B-cell linker |
| chr21_+_33671264 | 0.21 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr9_-_5830768 | 0.21 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr12_-_21810765 | 0.20 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr19_+_39390320 | 0.20 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr12_-_67197760 | 0.20 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr2_+_152266604 | 0.20 |
ENST00000430328.2
|
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr3_+_172361483 | 0.20 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr5_-_146889619 | 0.20 |
ENST00000343218.5
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr1_-_153518270 | 0.20 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr1_+_228337553 | 0.19 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chrX_+_134478706 | 0.19 |
ENST00000370761.3
ENST00000339249.4 ENST00000370760.3 |
ZNF449
|
zinc finger protein 449 |
| chr8_-_80680078 | 0.19 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr15_+_89182178 | 0.19 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr13_+_95364963 | 0.19 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chrX_+_153170455 | 0.19 |
ENST00000430697.1
ENST00000337474.5 ENST00000370049.1 |
AVPR2
|
arginine vasopressin receptor 2 |
| chr11_-_88796803 | 0.19 |
ENST00000418177.2
ENST00000455756.2 |
GRM5
|
glutamate receptor, metabotropic 5 |
| chr21_+_45432174 | 0.19 |
ENST00000380221.3
ENST00000291574.4 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr18_+_42260861 | 0.19 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr8_-_94752946 | 0.19 |
ENST00000519109.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr11_-_65325430 | 0.19 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chrX_+_114423963 | 0.18 |
ENST00000424776.3
|
RBMXL3
|
RNA binding motif protein, X-linked-like 3 |
| chrX_+_103029314 | 0.18 |
ENST00000429977.1
|
PLP1
|
proteolipid protein 1 |
| chr1_-_152595579 | 0.17 |
ENST00000335674.1
|
LCE3A
|
late cornified envelope 3A |
| chr19_-_46234119 | 0.17 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr9_-_7800067 | 0.17 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chr2_-_230787879 | 0.16 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr1_-_153517473 | 0.16 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr1_+_202431859 | 0.16 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr4_-_123377880 | 0.16 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr1_-_62190793 | 0.16 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr3_+_4535155 | 0.15 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chrX_+_136648643 | 0.14 |
ENST00000370606.3
|
ZIC3
|
Zic family member 3 |
| chr18_+_33877654 | 0.14 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr5_+_134094461 | 0.14 |
ENST00000452510.2
ENST00000354283.4 |
DDX46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr1_-_33786699 | 0.14 |
ENST00000442999.3
|
A3GALT2
|
alpha 1,3-galactosyltransferase 2 |
| chr1_-_38397384 | 0.13 |
ENST00000373027.1
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa |
| chr6_-_27841289 | 0.13 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr8_-_28747424 | 0.13 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr3_+_187930429 | 0.13 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_175870085 | 0.13 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr16_-_29757272 | 0.13 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chrY_-_25345070 | 0.13 |
ENST00000382510.4
ENST00000426000.2 ENST00000540248.1 ENST00000405239.1 |
DAZ1
|
deleted in azoospermia 1 |
| chr19_+_751122 | 0.13 |
ENST00000215582.6
|
MISP
|
mitotic spindle positioning |
| chr12_+_6961279 | 0.12 |
ENST00000229268.8
ENST00000389231.5 ENST00000542087.1 |
USP5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr12_+_50479101 | 0.12 |
ENST00000551966.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr16_-_58663720 | 0.11 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr12_+_25205568 | 0.11 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr1_-_202679535 | 0.11 |
ENST00000367268.4
|
SYT2
|
synaptotagmin II |
| chr10_-_7708918 | 0.11 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr11_-_65325203 | 0.11 |
ENST00000526927.1
ENST00000536982.1 |
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr7_+_99156145 | 0.11 |
ENST00000452314.1
ENST00000252713.4 |
ZNF655
|
zinc finger protein 655 |
| chr1_-_25291475 | 0.10 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0060262 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 1.3 | 5.2 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.9 | 6.4 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.7 | 2.8 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.3 | 1.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.3 | 6.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.7 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.2 | 1.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 0.7 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.2 | 2.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 3.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 3.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.4 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.1 | 0.9 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 1.0 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.4 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.3 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.1 | 1.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.8 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.7 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 0.8 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.1 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.5 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.3 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 4.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.8 | GO:0060539 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.4 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.6 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.5 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 1.5 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.7 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.8 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 1.5 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.4 | 3.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 3.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.2 | 0.9 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 6.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.7 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.7 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 1.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 2.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 2.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 8.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 11.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.7 | 5.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.7 | 2.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.6 | 6.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 1.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 0.7 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.2 | 6.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 0.8 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 1.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.7 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 1.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 2.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 3.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.4 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 1.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.8 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 2.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 1.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.7 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 6.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.9 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 5.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 2.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |