Project
Illumina Body Map 2, young vs old
Navigation
Downloads
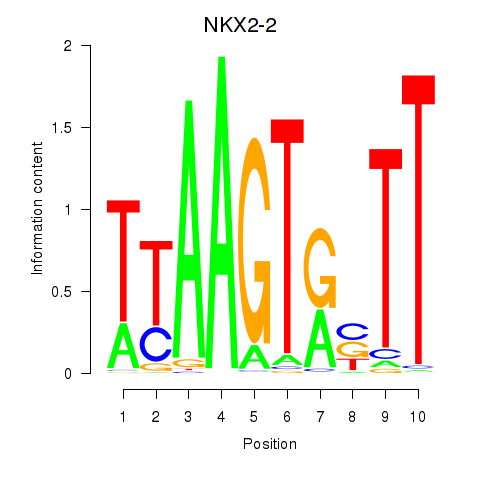
Results for NKX2-2
Z-value: 0.39
Transcription factors associated with NKX2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-2
|
ENSG00000125820.5 | NK2 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-2 | hg19_v2_chr20_-_21494654_21494678 | -0.33 | 6.5e-02 | Click! |
Activity profile of NKX2-2 motif
Sorted Z-values of NKX2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_54689532 | 2.82 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr7_+_37723450 | 2.61 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr7_+_37723336 | 2.52 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr9_-_85882145 | 1.86 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr3_-_150920979 | 1.70 |
ENST00000309180.5
ENST00000480322.1 |
GPR171
|
G protein-coupled receptor 171 |
| chr8_-_133772870 | 1.54 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr3_-_33686925 | 1.54 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_+_82767583 | 1.43 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr4_-_40477766 | 1.38 |
ENST00000507180.1
|
RBM47
|
RNA binding motif protein 47 |
| chr8_-_133772794 | 1.37 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr12_+_8666126 | 1.37 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr10_-_36813162 | 1.33 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr3_+_151591422 | 1.26 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr1_-_153363452 | 1.26 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr2_-_214013353 | 1.25 |
ENST00000451136.2
ENST00000421754.2 ENST00000374327.4 ENST00000413091.3 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_+_160203650 | 1.24 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr10_-_36812323 | 1.24 |
ENST00000543053.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chrM_+_12331 | 1.21 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_+_82767487 | 1.19 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr10_-_104913367 | 1.17 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr6_-_116833500 | 1.17 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr7_+_16700806 | 1.17 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr11_+_65266507 | 1.13 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr7_+_80253387 | 1.10 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr5_+_154181816 | 1.09 |
ENST00000518677.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr18_+_32558380 | 1.08 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr3_+_69811858 | 1.07 |
ENST00000433517.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr14_+_39734482 | 1.07 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chrX_-_15333775 | 1.06 |
ENST00000480796.1
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_+_102953608 | 1.03 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr7_+_80254279 | 1.02 |
ENST00000436384.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr4_+_95376396 | 1.02 |
ENST00000508216.1
ENST00000514743.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr21_+_30672433 | 1.02 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr15_-_60683326 | 1.01 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr12_+_69202795 | 0.99 |
ENST00000539479.1
ENST00000393415.3 ENST00000523991.1 ENST00000543323.1 ENST00000393416.2 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr1_+_84629976 | 0.98 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_-_154150651 | 0.97 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr5_+_75904918 | 0.96 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr12_+_69202975 | 0.95 |
ENST00000544561.1
ENST00000393410.1 ENST00000299252.4 ENST00000360430.2 ENST00000517852.1 ENST00000545204.1 ENST00000393413.3 ENST00000350057.5 ENST00000348801.2 ENST00000478070.1 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr2_-_145188137 | 0.95 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_148206156 | 0.94 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr22_-_50523807 | 0.93 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr17_-_29641084 | 0.93 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr22_-_43567750 | 0.91 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr7_-_24957699 | 0.91 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr14_-_89878369 | 0.91 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr3_+_63953415 | 0.90 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr16_+_53241854 | 0.90 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr3_-_33686743 | 0.90 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr13_-_45048386 | 0.89 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr7_-_142120321 | 0.87 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chr3_+_152017181 | 0.87 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr9_-_139361503 | 0.86 |
ENST00000453963.1
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr9_+_129097520 | 0.86 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr10_-_61513201 | 0.85 |
ENST00000414264.1
ENST00000594536.1 |
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr2_-_214015111 | 0.85 |
ENST00000433134.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr19_-_14889349 | 0.84 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr1_+_84630352 | 0.84 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr6_+_140175987 | 0.84 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr3_-_152058779 | 0.84 |
ENST00000408960.3
|
TMEM14E
|
transmembrane protein 14E |
| chr7_+_133615169 | 0.83 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr3_-_150966902 | 0.83 |
ENST00000424796.2
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr8_-_134072593 | 0.83 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr22_-_50523688 | 0.82 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr1_-_54405773 | 0.82 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chrX_-_70474910 | 0.82 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chrX_-_19817869 | 0.81 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chrX_+_123095890 | 0.81 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr20_-_60573188 | 0.81 |
ENST00000474089.1
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr21_+_17214724 | 0.81 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr17_+_66511224 | 0.81 |
ENST00000588178.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_+_84630053 | 0.81 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_+_39703084 | 0.80 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr1_-_216978709 | 0.80 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr1_+_104293028 | 0.80 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr3_-_69249863 | 0.80 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr5_+_75904950 | 0.79 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr3_+_142838091 | 0.78 |
ENST00000309575.3
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr3_+_41241596 | 0.78 |
ENST00000450969.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr18_-_67615088 | 0.77 |
ENST00000582621.1
|
CD226
|
CD226 molecule |
| chr20_+_30102231 | 0.77 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr14_+_61449076 | 0.77 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr11_+_64949899 | 0.77 |
ENST00000531068.1
ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr19_-_44174305 | 0.77 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr1_-_223308098 | 0.76 |
ENST00000342210.6
|
TLR5
|
toll-like receptor 5 |
| chr10_-_121296045 | 0.76 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr2_+_219110149 | 0.76 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr3_-_121379739 | 0.76 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr2_+_201994569 | 0.75 |
ENST00000457277.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_+_7445850 | 0.75 |
ENST00000593531.1
|
CTD-2207O23.3
|
Rho guanine nucleotide exchange factor 18 |
| chr9_+_131447342 | 0.75 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chrX_+_105445717 | 0.74 |
ENST00000372552.1
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr19_-_44174330 | 0.74 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr22_-_50523760 | 0.74 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chrX_-_154033661 | 0.74 |
ENST00000393531.1
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr1_-_150944411 | 0.73 |
ENST00000368949.4
|
CERS2
|
ceramide synthase 2 |
| chrX_-_70474499 | 0.73 |
ENST00000353904.2
|
ZMYM3
|
zinc finger, MYM-type 3 |
| chr3_+_98482175 | 0.73 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr2_+_74229812 | 0.72 |
ENST00000305799.7
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr2_-_214014959 | 0.72 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr2_-_38056538 | 0.72 |
ENST00000413792.1
|
LINC00211
|
long intergenic non-protein coding RNA 211 |
| chrX_+_123095860 | 0.71 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr10_-_94003003 | 0.71 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_-_58240470 | 0.71 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr4_+_89299885 | 0.71 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr3_+_190281229 | 0.70 |
ENST00000453359.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr5_+_179135246 | 0.70 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr12_-_49582978 | 0.70 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr5_-_78281775 | 0.69 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr14_-_31856397 | 0.69 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr7_+_115862858 | 0.68 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr5_-_88180342 | 0.68 |
ENST00000502983.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr9_-_20622478 | 0.68 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chrX_+_123095546 | 0.68 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr6_+_158733692 | 0.67 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr11_-_13517565 | 0.67 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr1_-_207095324 | 0.67 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr12_-_70093111 | 0.67 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr13_-_41240717 | 0.67 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr5_-_58652788 | 0.67 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_42631529 | 0.66 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr12_+_9066472 | 0.66 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr1_-_155880672 | 0.65 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr11_-_11374904 | 0.65 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr14_+_23654525 | 0.65 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr2_+_224822121 | 0.65 |
ENST00000258383.3
|
MRPL44
|
mitochondrial ribosomal protein L44 |
| chr14_+_22520762 | 0.64 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr2_-_145275828 | 0.64 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_135394840 | 0.64 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr1_+_117963209 | 0.64 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr2_+_201994208 | 0.64 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr9_+_137218362 | 0.63 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr1_-_31538517 | 0.63 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chrX_-_15333736 | 0.63 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr17_-_29641104 | 0.63 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr14_-_100841794 | 0.62 |
ENST00000556295.1
ENST00000554820.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr7_+_2393714 | 0.61 |
ENST00000431643.1
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr1_+_84630367 | 0.61 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_+_98741041 | 0.61 |
ENST00000286067.2
|
C10orf12
|
chromosome 10 open reading frame 12 |
| chr22_-_42086477 | 0.61 |
ENST00000402458.1
|
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr17_+_66521936 | 0.61 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr6_+_10633993 | 0.60 |
ENST00000417671.1
|
GCNT6
|
glucosaminyl (N-acetyl) transferase 6 |
| chr9_+_103340354 | 0.60 |
ENST00000307584.5
|
MURC
|
muscle-related coiled-coil protein |
| chr1_-_207095212 | 0.60 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr12_+_32552451 | 0.60 |
ENST00000534526.2
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr11_+_93479588 | 0.60 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr13_+_111855399 | 0.60 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr2_-_27558270 | 0.59 |
ENST00000454704.1
|
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr12_-_10978957 | 0.59 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr6_+_159084188 | 0.59 |
ENST00000367081.3
|
SYTL3
|
synaptotagmin-like 3 |
| chrX_-_23761317 | 0.59 |
ENST00000492081.1
ENST00000379303.5 ENST00000336430.7 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr3_-_113897899 | 0.59 |
ENST00000383673.2
ENST00000295881.7 |
DRD3
|
dopamine receptor D3 |
| chr6_-_150067632 | 0.57 |
ENST00000460354.2
ENST00000367404.4 ENST00000543637.1 |
NUP43
|
nucleoporin 43kDa |
| chr1_+_161691353 | 0.57 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr7_-_38339890 | 0.57 |
ENST00000390341.2
|
TRGV10
|
T cell receptor gamma variable 10 (non-functional) |
| chr2_+_183582774 | 0.56 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr5_+_118812294 | 0.55 |
ENST00000509514.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr8_-_101962777 | 0.55 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr10_-_27529486 | 0.55 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr3_+_98482159 | 0.55 |
ENST00000468553.1
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr6_+_20534672 | 0.55 |
ENST00000274695.4
ENST00000378624.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr13_+_113863858 | 0.54 |
ENST00000375440.4
|
CUL4A
|
cullin 4A |
| chr3_-_196911002 | 0.54 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr9_+_116263639 | 0.54 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr5_-_58335281 | 0.54 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr13_+_111855414 | 0.54 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr10_+_63808970 | 0.53 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr9_+_129097479 | 0.53 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr1_+_54359854 | 0.53 |
ENST00000361921.3
ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1
|
deiodinase, iodothyronine, type I |
| chr17_+_7591747 | 0.53 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr20_-_36152914 | 0.53 |
ENST00000397131.1
|
BLCAP
|
bladder cancer associated protein |
| chr13_+_25875785 | 0.53 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr9_-_86593238 | 0.53 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr15_+_79165296 | 0.53 |
ENST00000558746.1
ENST00000558830.1 ENST00000559345.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr14_-_100841670 | 0.52 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr7_-_944631 | 0.52 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr15_+_81391740 | 0.52 |
ENST00000561216.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr4_+_87928140 | 0.52 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr18_-_43684230 | 0.52 |
ENST00000592989.1
ENST00000589869.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr10_+_81462983 | 0.52 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr18_-_60986613 | 0.51 |
ENST00000444484.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr15_-_56209306 | 0.51 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr8_-_124279627 | 0.51 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr15_-_60690163 | 0.51 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr14_-_100841930 | 0.51 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr15_+_79165372 | 0.50 |
ENST00000558502.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr13_-_99630233 | 0.50 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr5_+_118812237 | 0.50 |
ENST00000513628.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr17_+_7591639 | 0.50 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr1_-_9953295 | 0.50 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr6_+_148593425 | 0.50 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr11_+_94277017 | 0.50 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr14_+_22964877 | 0.50 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr5_-_58571935 | 0.50 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr22_+_42086547 | 0.50 |
ENST00000402966.1
|
C22orf46
|
chromosome 22 open reading frame 46 |
| chr12_-_10875831 | 0.49 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr20_+_30697510 | 0.49 |
ENST00000217315.5
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr18_-_60985914 | 0.49 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr12_+_14572070 | 0.49 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_162087577 | 0.49 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.4 | 1.5 | GO:0044145 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.3 | 1.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.3 | 1.0 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.3 | 3.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 0.9 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.3 | 1.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 2.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 0.8 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.3 | 0.5 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.3 | 0.8 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.3 | 1.0 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.2 | 1.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.7 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.2 | 0.7 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 0.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 0.7 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 0.6 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 1.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 1.4 | GO:0019075 | virus maturation(GO:0019075) |
| 0.2 | 0.8 | GO:1904499 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.2 | 2.1 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 0.7 | GO:1905044 | Schwann cell proliferation involved in axon regeneration(GO:0014011) negative regulation of Schwann cell migration(GO:1900148) regulation of Schwann cell proliferation involved in axon regeneration(GO:1905044) negative regulation of Schwann cell proliferation involved in axon regeneration(GO:1905045) |
| 0.2 | 1.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.2 | 0.5 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 1.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.2 | 1.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.2 | 1.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 2.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.6 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 2.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 3.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.8 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 0.8 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 1.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.5 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.5 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.7 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.7 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.4 | GO:0042369 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.6 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.3 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 2.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 2.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.1 | 0.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 1.5 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.4 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 1.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.3 | GO:0061569 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.4 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.2 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.3 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.2 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.1 | 0.5 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.4 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.7 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 0.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 1.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.9 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 1.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.3 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.6 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.3 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 0.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.6 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 1.3 | GO:0002544 | leukocyte migration involved in inflammatory response(GO:0002523) chronic inflammatory response(GO:0002544) |
| 0.0 | 0.8 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.8 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.5 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 1.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.5 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.5 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.3 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.8 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.8 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 1.6 | GO:2000142 | regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 1.3 | GO:0097503 | cellular response to interleukin-6(GO:0071354) sialylation(GO:0097503) |
| 0.0 | 0.4 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.7 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.6 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.5 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 1.0 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.1 | GO:1901529 | transepithelial water transport(GO:0035377) positive regulation of anion channel activity(GO:1901529) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) positive regulation of inorganic anion transmembrane transport(GO:1903797) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.4 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 1.2 | GO:0016569 | covalent chromatin modification(GO:0016569) |
| 0.0 | 0.2 | GO:0090050 | peptidyl-lysine deacetylation(GO:0034983) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.7 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.5 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.3 | 0.8 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 0.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 0.8 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.2 | 4.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.2 | 2.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.7 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 1.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.8 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 1.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.5 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.5 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 4.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.0 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.4 | 1.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.3 | 1.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 0.9 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.3 | 1.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 1.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 1.0 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.2 | 0.7 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.2 | 0.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.2 | 0.8 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 2.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 1.1 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.2 | 0.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 0.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 3.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.6 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.4 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 1.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 2.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 2.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 2.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.3 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 1.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 2.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 4.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 1.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.8 | GO:0017162 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 8.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 1.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.1 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 3.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.7 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 2.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.0 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |