Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for NKX2-6
Z-value: 0.56
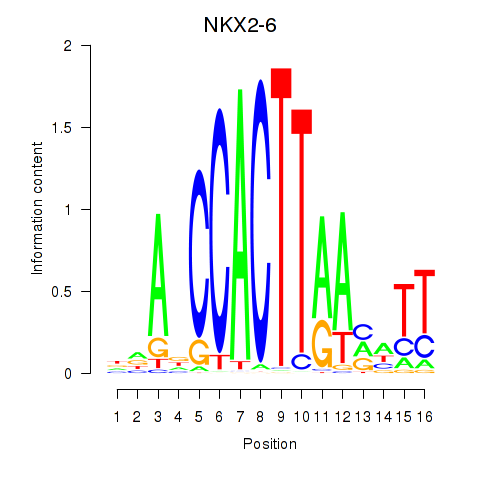
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.6 | NK2 homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-6 | hg19_v2_chr8_-_23563922_23564111 | -0.46 | 8.8e-03 | Click! |
Activity profile of NKX2-6 motif
Sorted Z-values of NKX2-6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22788560 | 6.31 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr19_+_3178736 | 4.50 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr22_-_37882395 | 4.42 |
ENST00000416983.3
ENST00000424765.2 ENST00000356998.3 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_+_142498725 | 4.29 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr7_+_142028105 | 4.02 |
ENST00000390353.2
|
TRBV6-1
|
T cell receptor beta variable 6-1 |
| chr14_+_22580233 | 3.90 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr22_+_22723969 | 3.86 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr6_-_159466136 | 3.82 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr9_-_137809718 | 3.76 |
ENST00000371806.3
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1 |
| chr6_-_159466042 | 3.48 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr22_+_44577237 | 3.47 |
ENST00000415224.1
ENST00000417767.1 |
PARVG
|
parvin, gamma |
| chr12_-_15103621 | 3.37 |
ENST00000536592.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_158900568 | 3.34 |
ENST00000458222.1
|
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr22_+_23229960 | 3.18 |
ENST00000526893.1
ENST00000532223.2 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda-like polypeptide 5 |
| chr7_-_38389573 | 3.16 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5 |
| chr12_+_113354341 | 3.14 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_-_121379739 | 2.97 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr3_-_120365866 | 2.94 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr1_+_158259558 | 2.91 |
ENST00000368170.3
|
CD1C
|
CD1c molecule |
| chr3_+_114012819 | 2.89 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr15_+_81475047 | 2.87 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr2_+_219246746 | 2.85 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr17_-_29645836 | 2.84 |
ENST00000578584.1
|
CTD-2370N5.3
|
CTD-2370N5.3 |
| chr14_+_22265444 | 2.81 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr15_-_55563072 | 2.81 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr22_-_24096562 | 2.79 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr22_+_22749343 | 2.69 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr2_-_87088995 | 2.66 |
ENST00000393759.2
ENST00000349455.3 ENST00000331469.2 ENST00000431506.2 ENST00000393761.2 ENST00000390655.6 |
CD8B
|
CD8b molecule |
| chr16_-_20556492 | 2.56 |
ENST00000568098.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr9_+_215158 | 2.54 |
ENST00000479404.1
|
DOCK8
|
dedicator of cytokinesis 8 |
| chr2_-_182521823 | 2.52 |
ENST00000410087.3
ENST00000409440.3 |
CERKL
|
ceramide kinase-like |
| chr4_-_48082192 | 2.47 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr1_-_108231101 | 2.43 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr22_-_24096630 | 2.42 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr14_+_91709103 | 2.38 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr1_+_158223923 | 2.25 |
ENST00000289429.5
|
CD1A
|
CD1a molecule |
| chr12_-_68845165 | 2.19 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr2_-_112237835 | 2.15 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr16_-_20367584 | 2.09 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr11_-_65629497 | 2.08 |
ENST00000532134.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr11_-_104827425 | 2.08 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr12_+_14561422 | 2.05 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr6_-_32977345 | 2.04 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr14_+_23002427 | 2.01 |
ENST00000390527.1
|
TRAJ10
|
T cell receptor alpha joining 10 |
| chrY_+_15418467 | 2.01 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr19_+_7701985 | 2.00 |
ENST00000595950.1
ENST00000441779.2 ENST00000221283.5 ENST00000414284.2 |
STXBP2
|
syntaxin binding protein 2 |
| chr14_-_60337684 | 1.99 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr17_-_57229155 | 1.96 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr14_-_106805716 | 1.89 |
ENST00000438142.2
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr19_-_43835582 | 1.86 |
ENST00000595748.1
|
CTC-490G23.2
|
CTC-490G23.2 |
| chr22_+_44577724 | 1.81 |
ENST00000466375.2
|
PARVG
|
parvin, gamma |
| chr13_-_30951074 | 1.79 |
ENST00000416261.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr12_-_371994 | 1.78 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr2_+_87769459 | 1.76 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr9_-_4859260 | 1.73 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr2_+_102927962 | 1.72 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr19_-_49843539 | 1.71 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr17_-_72527605 | 1.66 |
ENST00000392621.1
ENST00000314401.3 |
CD300LB
|
CD300 molecule-like family member b |
| chr3_-_46249878 | 1.64 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr9_+_125132803 | 1.63 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr11_+_122709200 | 1.62 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr11_-_6640585 | 1.58 |
ENST00000533371.1
ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1
|
tripeptidyl peptidase I |
| chr11_-_65624415 | 1.57 |
ENST00000524553.1
ENST00000527344.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr3_+_121796697 | 1.57 |
ENST00000482356.1
ENST00000393627.2 |
CD86
|
CD86 molecule |
| chr13_-_30951282 | 1.53 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr12_-_89919965 | 1.52 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr19_+_55385682 | 1.50 |
ENST00000391726.3
|
FCAR
|
Fc fragment of IgA, receptor for |
| chr17_+_57807062 | 1.50 |
ENST00000587259.1
|
VMP1
|
vacuole membrane protein 1 |
| chr2_+_70142189 | 1.49 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr3_+_42544084 | 1.45 |
ENST00000543411.1
ENST00000438259.2 ENST00000439731.1 ENST00000325123.4 |
VIPR1
|
vasoactive intestinal peptide receptor 1 |
| chr9_+_127023704 | 1.38 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr19_-_44174305 | 1.38 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr6_+_33043703 | 1.36 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr11_-_62521614 | 1.35 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr9_+_125133315 | 1.33 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_-_89920030 | 1.32 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr10_-_6019552 | 1.31 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr3_+_186435137 | 1.31 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr12_-_39300071 | 1.30 |
ENST00000550863.1
|
CPNE8
|
copine VIII |
| chr10_-_6019984 | 1.28 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
| chr11_-_104480019 | 1.27 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr5_-_169725231 | 1.26 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr6_-_26216872 | 1.25 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr19_-_44174330 | 1.21 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr9_-_215744 | 1.21 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr1_+_16767167 | 1.20 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr7_+_31726822 | 1.19 |
ENST00000409146.3
|
PPP1R17
|
protein phosphatase 1, regulatory subunit 17 |
| chr5_-_111091948 | 1.16 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr4_+_89299885 | 1.16 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr10_+_121652204 | 1.16 |
ENST00000369075.3
ENST00000543134.1 |
SEC23IP
|
SEC23 interacting protein |
| chr10_-_6019455 | 1.14 |
ENST00000530685.1
ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr3_+_186435065 | 1.14 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr1_+_110162448 | 1.11 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr8_-_17942432 | 1.10 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr12_+_60083118 | 1.10 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_-_89919765 | 1.09 |
ENST00000541909.1
ENST00000313546.3 |
POC1B
|
POC1 centriolar protein B |
| chr1_-_21113105 | 1.09 |
ENST00000375000.1
ENST00000419490.1 ENST00000414993.1 ENST00000443615.1 ENST00000312239.5 |
HP1BP3
|
heterochromatin protein 1, binding protein 3 |
| chr17_+_41150793 | 1.07 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr17_+_41150479 | 1.06 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr2_-_100721801 | 1.06 |
ENST00000424600.1
ENST00000441400.1 |
AFF3
|
AF4/FMR2 family, member 3 |
| chr1_+_171217677 | 1.06 |
ENST00000402921.2
|
FMO1
|
flavin containing monooxygenase 1 |
| chr1_-_31538517 | 1.02 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr2_+_70142232 | 0.98 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr15_-_20170354 | 0.98 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr2_-_175462456 | 0.97 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr22_-_50523807 | 0.97 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr1_+_110163202 | 0.97 |
ENST00000531203.1
ENST00000256578.3 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr22_-_50523688 | 0.97 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr20_-_1373726 | 0.96 |
ENST00000400137.4
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr2_-_100721923 | 0.95 |
ENST00000356421.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr5_-_139937895 | 0.95 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr8_+_41386761 | 0.95 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr1_+_16767195 | 0.95 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr2_-_100721178 | 0.95 |
ENST00000409236.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr9_+_131452239 | 0.94 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr16_-_47493041 | 0.94 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr3_+_119499331 | 0.94 |
ENST00000393716.2
ENST00000466380.1 |
NR1I2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr2_-_202298268 | 0.93 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr22_-_50523843 | 0.93 |
ENST00000535444.1
ENST00000431262.2 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr6_+_42584847 | 0.91 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr11_+_118754475 | 0.91 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr5_-_18746250 | 0.90 |
ENST00000512978.1
|
CTD-2023M8.1
|
CTD-2023M8.1 |
| chr10_-_101841588 | 0.90 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr19_-_2282167 | 0.88 |
ENST00000342063.3
|
C19orf35
|
chromosome 19 open reading frame 35 |
| chr21_-_46964306 | 0.87 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.1 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr11_-_129817448 | 0.86 |
ENST00000304538.6
|
PRDM10
|
PR domain containing 10 |
| chr8_+_20054878 | 0.84 |
ENST00000276390.2
ENST00000519667.1 |
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
| chr9_+_116263639 | 0.83 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr20_-_1373682 | 0.82 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr17_-_34890037 | 0.81 |
ENST00000589404.1
|
MYO19
|
myosin XIX |
| chr2_-_25565377 | 0.81 |
ENST00000264709.3
ENST00000406659.3 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr17_+_41150290 | 0.80 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr2_-_175462934 | 0.80 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr14_+_65182395 | 0.79 |
ENST00000554088.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr9_+_4839762 | 0.79 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr9_-_19065082 | 0.79 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr11_-_129817356 | 0.78 |
ENST00000526082.1
|
PRDM10
|
PR domain containing 10 |
| chr8_-_145086922 | 0.78 |
ENST00000530478.1
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr9_-_70465758 | 0.77 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr17_-_6524159 | 0.76 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr11_+_94277017 | 0.76 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr4_+_70916119 | 0.76 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr3_-_39196049 | 0.74 |
ENST00000514182.1
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr17_-_7216939 | 0.73 |
ENST00000573684.1
|
GPS2
|
G protein pathway suppressor 2 |
| chr6_+_10528560 | 0.73 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr15_-_65579177 | 0.73 |
ENST00000444347.2
ENST00000261888.6 |
PARP16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr1_-_1763721 | 0.73 |
ENST00000437146.1
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr4_+_17579110 | 0.72 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr20_-_1373606 | 0.72 |
ENST00000381715.1
ENST00000439640.2 ENST00000381719.3 |
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr22_-_50523760 | 0.69 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr1_+_158969752 | 0.68 |
ENST00000566111.1
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr3_-_42003613 | 0.67 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr3_-_56950407 | 0.67 |
ENST00000496106.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr17_-_29624343 | 0.67 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr11_+_65627865 | 0.65 |
ENST00000308110.4
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr12_+_118454500 | 0.65 |
ENST00000537315.1
ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5
|
replication factor C (activator 1) 5, 36.5kDa |
| chr4_-_170924888 | 0.64 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chrX_-_47341928 | 0.64 |
ENST00000313116.7
|
ZNF41
|
zinc finger protein 41 |
| chr2_-_31361543 | 0.63 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chrX_+_139791917 | 0.63 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr3_+_171561127 | 0.62 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr1_-_78444738 | 0.62 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr1_+_173793777 | 0.62 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr17_+_66511224 | 0.62 |
ENST00000588178.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_+_1243947 | 0.61 |
ENST00000379031.5
|
PUSL1
|
pseudouridylate synthase-like 1 |
| chr4_+_91048706 | 0.61 |
ENST00000509176.1
|
CCSER1
|
coiled-coil serine-rich protein 1 |
| chr17_-_5522731 | 0.60 |
ENST00000576905.1
|
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr1_+_159796534 | 0.60 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr1_-_149400542 | 0.60 |
ENST00000392948.2
|
HIST2H3PS2
|
histone cluster 2, H3, pseudogene 2 |
| chr7_+_141463897 | 0.59 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr13_-_26452679 | 0.59 |
ENST00000596729.1
|
AL138815.2
|
Uncharacterized protein |
| chr17_-_34890665 | 0.58 |
ENST00000586007.1
|
MYO19
|
myosin XIX |
| chr1_+_54569968 | 0.58 |
ENST00000391366.1
|
AL161915.1
|
Uncharacterized protein |
| chrX_+_52841228 | 0.57 |
ENST00000351072.1
ENST00000425386.1 ENST00000375503.3 |
XAGE5
|
X antigen family, member 5 |
| chr3_-_48732926 | 0.57 |
ENST00000453202.1
|
IP6K2
|
inositol hexakisphosphate kinase 2 |
| chr6_-_34664612 | 0.56 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr5_-_133510456 | 0.56 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chrX_-_47518498 | 0.54 |
ENST00000335890.2
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr17_+_33914424 | 0.53 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr7_+_75544466 | 0.53 |
ENST00000421059.1
ENST00000394893.1 ENST00000412521.1 ENST00000414186.1 |
POR
|
P450 (cytochrome) oxidoreductase |
| chr17_-_34890732 | 0.53 |
ENST00000268852.9
|
MYO19
|
myosin XIX |
| chr1_-_115259337 | 0.52 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr1_+_159796589 | 0.52 |
ENST00000368104.4
|
SLAMF8
|
SLAM family member 8 |
| chr2_-_69870747 | 0.52 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr3_-_187694195 | 0.52 |
ENST00000446091.1
|
RP11-132N15.3
|
RP11-132N15.3 |
| chr1_+_156338993 | 0.52 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr19_+_58144529 | 0.52 |
ENST00000347302.3
ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211
|
zinc finger protein 211 |
| chr4_+_70146217 | 0.51 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chrX_-_47518527 | 0.50 |
ENST00000333119.3
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr6_+_126102292 | 0.49 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr11_+_65627974 | 0.49 |
ENST00000525768.1
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr17_-_34890617 | 0.48 |
ENST00000586886.1
ENST00000585719.1 ENST00000585818.1 |
MYO19
|
myosin XIX |
| chr6_-_15586238 | 0.47 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr1_-_173793246 | 0.47 |
ENST00000345664.6
ENST00000367710.3 |
CENPL
|
centromere protein L |
| chr10_+_135204338 | 0.46 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr17_+_25621102 | 0.45 |
ENST00000581440.1
ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1
|
WD repeat and SOCS box containing 1 |
| chr3_-_160167301 | 0.45 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chr5_+_52083730 | 0.45 |
ENST00000282588.6
ENST00000274311.2 |
ITGA1
PELO
|
integrin, alpha 1 pelota homolog (Drosophila) |
| chr19_+_17186577 | 0.44 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr19_-_51611623 | 0.44 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.9 | 2.8 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.8 | 2.5 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.7 | 2.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.5 | 3.7 | GO:0051510 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.5 | 1.6 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.5 | 3.4 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.4 | 5.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 2.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 2.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.4 | 2.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 2.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 2.6 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 0.9 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.3 | 1.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 4.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 1.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 2.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 0.9 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 4.5 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 0.6 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.2 | 2.5 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.2 | 2.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 2.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 0.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 0.5 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) |
| 0.2 | 1.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.5 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 3.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 3.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 3.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.6 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.1 | 0.8 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 2.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.3 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.9 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 8.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 2.9 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.1 | 0.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.6 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 1.8 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.4 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 1.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 3.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 2.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.8 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.4 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 2.0 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.9 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 2.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 2.4 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 9.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 3.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 1.0 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 1.4 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.7 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 1.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 1.0 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.1 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.5 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 1.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 1.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.3 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 4.5 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 3.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 1.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 1.5 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.8 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 3.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 2.0 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 2.5 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 1.4 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 3.0 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.7 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 1.0 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 7.8 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.0 | GO:2000466 | negative regulation of glycogen (starch) synthase activity(GO:2000466) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.4 | 3.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 1.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 0.9 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 2.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 2.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 0.6 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 1.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 8.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 3.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 0.3 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 2.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 3.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 4.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 3.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 2.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 5.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.0 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 3.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 1.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 3.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 3.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 3.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 2.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 3.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 3.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 2.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 9.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 3.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 3.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.9 | 4.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.6 | 3.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 5.2 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.4 | 1.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.1 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.4 | 1.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 2.6 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.3 | 4.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 1.5 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.3 | 3.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 3.0 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.3 | 3.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 3.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 0.9 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.2 | 2.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 2.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 2.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 2.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 0.5 | GO:0047726 | iron-cytochrome-c reductase activity(GO:0047726) |
| 0.2 | 2.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 1.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.2 | 0.8 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.2 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.9 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 8.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 2.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.6 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 2.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 2.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.3 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.1 | GO:0001159 | core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 2.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 2.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 3.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.8 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.4 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.1 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 8.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.9 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 1.2 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 0.7 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 2.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 2.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 3.7 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 2.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 3.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 3.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 7.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.9 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 6.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 3.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 3.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 5.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 5.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 3.7 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 2.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 3.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 4.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 10.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 3.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 2.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 3.0 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 4.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 3.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |