Project
Illumina Body Map 2, young vs old
Navigation
Downloads
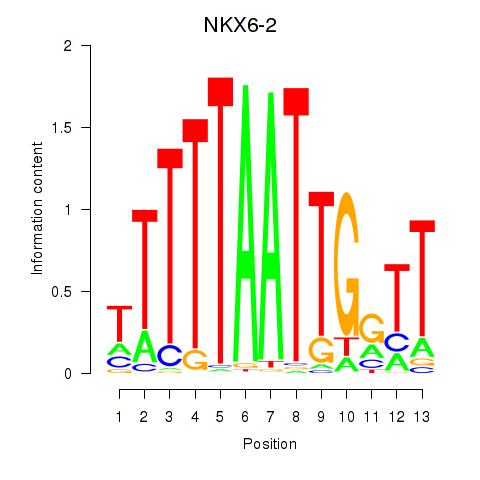
Results for NKX6-2
Z-value: 0.08
Transcription factors associated with NKX6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-2
|
ENSG00000148826.6 | NK6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX6-2 | hg19_v2_chr10_-_134599556_134599653 | 0.57 | 5.9e-04 | Click! |
Activity profile of NKX6-2 motif
Sorted Z-values of NKX6-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_39509070 | 2.61 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr12_+_41136144 | 2.60 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr4_+_41258786 | 2.58 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr1_+_110993795 | 2.56 |
ENST00000271331.3
|
PROK1
|
prokineticin 1 |
| chr17_-_42992856 | 2.18 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr3_-_79816965 | 2.15 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr14_-_60097524 | 2.15 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr3_+_39509163 | 2.15 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr1_+_170632250 | 1.96 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr9_+_109685630 | 1.91 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr6_+_12290586 | 1.90 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr2_+_108994633 | 1.89 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chrX_-_103087136 | 1.88 |
ENST00000243298.2
|
RAB9B
|
RAB9B, member RAS oncogene family |
| chr5_+_161275320 | 1.86 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr10_+_26505179 | 1.85 |
ENST00000376261.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr2_+_200472779 | 1.83 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr2_-_217560248 | 1.82 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr2_-_175711978 | 1.78 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr18_+_32290218 | 1.77 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr2_-_73520667 | 1.75 |
ENST00000545030.1
ENST00000436467.2 |
EGR4
|
early growth response 4 |
| chr2_-_217559517 | 1.73 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr2_+_210444142 | 1.73 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr11_-_126810521 | 1.69 |
ENST00000530572.1
|
RP11-688I9.4
|
RP11-688I9.4 |
| chr14_-_27066636 | 1.67 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr4_-_185139062 | 1.64 |
ENST00000296741.2
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr2_+_210444298 | 1.57 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chrX_+_36254051 | 1.56 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr14_+_101295638 | 1.55 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr2_-_175712479 | 1.52 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr2_+_210443993 | 1.52 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_-_175712270 | 1.51 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr4_-_87374283 | 1.47 |
ENST00000361569.2
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr14_+_72399833 | 1.45 |
ENST00000553530.1
ENST00000556437.1 |
RGS6
|
regulator of G-protein signaling 6 |
| chr9_+_82186872 | 1.41 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr14_-_60097297 | 1.39 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr1_-_114696472 | 1.37 |
ENST00000393296.1
ENST00000369547.1 ENST00000610222.1 |
SYT6
|
synaptotagmin VI |
| chr2_-_211342292 | 1.35 |
ENST00000448951.1
|
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr4_-_87374330 | 1.34 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_-_74686314 | 1.33 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr5_+_161495038 | 1.25 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr12_-_6233828 | 1.25 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr9_-_23825956 | 1.22 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr3_+_178276488 | 1.21 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr3_-_138048682 | 1.21 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr3_-_98241358 | 1.20 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr9_+_130026756 | 1.19 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr1_+_85527987 | 1.19 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr7_-_107880508 | 1.18 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr8_+_104831472 | 1.17 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_-_71169031 | 1.15 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr3_-_147124547 | 1.14 |
ENST00000491672.1
ENST00000383075.3 |
ZIC4
|
Zic family member 4 |
| chr8_+_24298531 | 1.14 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr12_-_28124903 | 1.14 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chrX_+_102840408 | 1.12 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr1_-_49242553 | 1.10 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr5_-_58295712 | 1.10 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_+_24298597 | 1.06 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr1_-_190446759 | 1.06 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr8_+_24298438 | 1.06 |
ENST00000441335.2
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr6_-_26043885 | 1.05 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr5_-_88120083 | 1.04 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr1_+_244515930 | 1.01 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr1_-_243326612 | 1.00 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr5_+_155753745 | 1.00 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr5_-_88120151 | 1.00 |
ENST00000506716.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chrX_-_138287168 | 1.00 |
ENST00000436198.1
|
FGF13
|
fibroblast growth factor 13 |
| chr17_-_57604227 | 0.98 |
ENST00000584262.1
|
RP11-567L7.6
|
RP11-567L7.6 |
| chr16_+_25228242 | 0.98 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr3_+_156799587 | 0.97 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr4_-_140201333 | 0.96 |
ENST00000398955.1
|
MGARP
|
mitochondria-localized glutamic acid-rich protein |
| chr2_-_80531399 | 0.96 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chrX_-_80457385 | 0.95 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr9_-_23826298 | 0.94 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr5_+_161274685 | 0.93 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr18_+_32402321 | 0.93 |
ENST00000587723.1
|
DTNA
|
dystrobrevin, alpha |
| chr3_+_16216210 | 0.92 |
ENST00000437509.1
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr5_+_161274940 | 0.92 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr21_-_36421626 | 0.92 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr5_+_166711804 | 0.91 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr14_-_27066960 | 0.90 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chrX_+_35937843 | 0.90 |
ENST00000297866.5
|
CXorf22
|
chromosome X open reading frame 22 |
| chr2_+_233527443 | 0.90 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chrX_+_135230712 | 0.88 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr15_+_49715449 | 0.86 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr22_+_17956618 | 0.86 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr1_-_109935819 | 0.86 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr5_-_20575959 | 0.83 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr21_-_36421535 | 0.83 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr15_-_96036999 | 0.83 |
ENST00000554412.2
|
RP11-398J10.2
|
RP11-398J10.2 |
| chr10_+_50887683 | 0.82 |
ENST00000374113.3
ENST00000374111.3 ENST00000374112.3 ENST00000535836.1 |
C10orf53
|
chromosome 10 open reading frame 53 |
| chr15_+_49715293 | 0.82 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr19_-_49220084 | 0.80 |
ENST00000595591.1
ENST00000356751.4 ENST00000594582.1 |
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr2_-_80531720 | 0.80 |
ENST00000416268.1
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr15_+_74421961 | 0.79 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr1_+_115397424 | 0.78 |
ENST00000369522.3
ENST00000455987.1 |
SYCP1
|
synaptonemal complex protein 1 |
| chr7_+_123241908 | 0.76 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr6_+_26199737 | 0.76 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr19_+_12175535 | 0.74 |
ENST00000550826.1
|
ZNF844
|
zinc finger protein 844 |
| chr18_+_28923589 | 0.72 |
ENST00000462981.2
|
DSG1
|
desmoglein 1 |
| chr16_+_70695570 | 0.71 |
ENST00000597002.1
|
FLJ00418
|
FLJ00418 |
| chr14_+_52164820 | 0.70 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr6_-_80247140 | 0.70 |
ENST00000392959.1
ENST00000467898.3 |
LCA5
|
Leber congenital amaurosis 5 |
| chr18_-_12656715 | 0.69 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr6_-_131277510 | 0.69 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr3_+_16216137 | 0.69 |
ENST00000339732.5
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chrX_+_10124977 | 0.69 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr3_+_159557637 | 0.68 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr2_+_108994466 | 0.68 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr6_-_56716686 | 0.67 |
ENST00000520645.1
|
DST
|
dystonin |
| chr9_-_16215897 | 0.66 |
ENST00000433347.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr9_-_93405352 | 0.66 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr20_+_57467204 | 0.65 |
ENST00000603546.1
|
GNAS
|
GNAS complex locus |
| chr1_-_247887345 | 0.65 |
ENST00000366485.1
|
OR14A2
|
olfactory receptor, family 14, subfamily A, member 2 |
| chr12_+_111284764 | 0.63 |
ENST00000545036.1
ENST00000308208.5 |
CCDC63
|
coiled-coil domain containing 63 |
| chr7_-_78400364 | 0.62 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chrX_+_135388147 | 0.61 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr10_+_123951957 | 0.61 |
ENST00000514539.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_-_80247105 | 0.59 |
ENST00000369846.4
|
LCA5
|
Leber congenital amaurosis 5 |
| chr8_+_79428539 | 0.59 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr10_+_32873190 | 0.59 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr15_-_89764929 | 0.58 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr3_-_193272874 | 0.57 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr15_-_42302445 | 0.57 |
ENST00000413860.2
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr8_-_82395461 | 0.57 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr9_-_21385396 | 0.57 |
ENST00000380206.2
|
IFNA2
|
interferon, alpha 2 |
| chr12_+_25205568 | 0.56 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr19_+_12175596 | 0.56 |
ENST00000441304.2
|
ZNF844
|
zinc finger protein 844 |
| chr18_-_52989217 | 0.55 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr4_+_26324474 | 0.54 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr8_+_104831554 | 0.54 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr14_-_101295407 | 0.54 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr3_+_140981456 | 0.52 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr6_+_97753454 | 0.52 |
ENST00000574739.1
|
RP3-418C23.2
|
RP3-418C23.2 |
| chr4_+_190580748 | 0.52 |
ENST00000513681.1
|
RP11-462G22.1
|
RP11-462G22.1 |
| chr1_+_151253991 | 0.51 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr1_-_21377447 | 0.51 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr3_-_45837959 | 0.51 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chrX_+_55744166 | 0.51 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr1_-_110933663 | 0.50 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr6_+_158733692 | 0.50 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr12_+_7022909 | 0.49 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr1_-_110933611 | 0.49 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr5_+_95066823 | 0.49 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr6_+_47666275 | 0.49 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr1_+_201617264 | 0.49 |
ENST00000367296.4
|
NAV1
|
neuron navigator 1 |
| chr8_-_42358742 | 0.48 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr7_-_22233442 | 0.48 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_+_169757750 | 0.48 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr22_-_40929812 | 0.48 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr14_-_25479811 | 0.48 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr1_-_186430222 | 0.47 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr3_-_45838011 | 0.47 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr13_-_45048386 | 0.46 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr1_+_201617450 | 0.46 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr18_-_19994830 | 0.46 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr1_+_32379174 | 0.46 |
ENST00000391369.1
|
AL136115.1
|
HCG2032337; PRO1848; Uncharacterized protein |
| chr12_-_109458820 | 0.45 |
ENST00000550490.1
|
SVOP
|
SV2 related protein homolog (rat) |
| chrX_+_55744228 | 0.45 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chrX_+_134887233 | 0.45 |
ENST00000443882.1
|
CT45A3
|
cancer/testis antigen family 45, member A3 |
| chr14_+_52164675 | 0.45 |
ENST00000555936.1
|
FRMD6
|
FERM domain containing 6 |
| chr18_-_52989525 | 0.44 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr1_+_109265033 | 0.43 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr14_+_88852059 | 0.43 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr17_-_67057047 | 0.43 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr7_-_137028534 | 0.42 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr3_+_68053359 | 0.42 |
ENST00000478136.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr2_+_179318295 | 0.42 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr6_-_33864684 | 0.41 |
ENST00000506222.2
ENST00000533304.1 |
LINC01016
|
long intergenic non-protein coding RNA 1016 |
| chr8_-_72268721 | 0.41 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr1_+_152178320 | 0.40 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr5_-_88119580 | 0.40 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr6_-_37665751 | 0.40 |
ENST00000297153.7
ENST00000434837.3 |
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr16_-_67517716 | 0.40 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr1_+_174933899 | 0.40 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr2_-_105882585 | 0.39 |
ENST00000595531.1
|
AC012360.2
|
LOC644617 protein; Uncharacterized protein |
| chr7_-_137028498 | 0.39 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr17_-_67057114 | 0.38 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr12_+_111284805 | 0.38 |
ENST00000552694.1
|
CCDC63
|
coiled-coil domain containing 63 |
| chr10_-_100995540 | 0.38 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr18_-_30716038 | 0.37 |
ENST00000581852.1
|
CCDC178
|
coiled-coil domain containing 178 |
| chr8_+_26150628 | 0.37 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr11_+_48510269 | 0.36 |
ENST00000446524.1
|
OR4A47
|
olfactory receptor, family 4, subfamily A, member 47 |
| chr5_-_146258205 | 0.36 |
ENST00000394413.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_16916624 | 0.35 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr9_+_88556036 | 0.35 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr7_-_84122033 | 0.35 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr8_+_104831440 | 0.35 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_112635675 | 0.35 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr21_-_36421401 | 0.34 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr10_+_91589309 | 0.34 |
ENST00000448490.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr2_+_179149636 | 0.34 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr11_-_9088284 | 0.34 |
ENST00000531429.1
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr17_-_67057203 | 0.34 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr12_-_53045948 | 0.34 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr12_+_54422142 | 0.33 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr7_-_78400598 | 0.33 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr8_-_15095832 | 0.33 |
ENST00000382080.1
|
SGCZ
|
sarcoglycan, zeta |
| chr16_+_14802801 | 0.32 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr14_-_75530693 | 0.32 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr4_-_108204846 | 0.32 |
ENST00000513208.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX6-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.9 | 2.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.6 | 1.7 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.5 | 1.9 | GO:0060585 | nitric oxide transport(GO:0030185) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.4 | 2.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.3 | 2.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 4.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.3 | 1.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 | 0.8 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.3 | 1.9 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.2 | 1.1 | GO:0035106 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.2 | 5.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 2.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 0.8 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 1.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 2.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 2.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.6 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 1.0 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 1.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 2.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 1.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.8 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 5.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.5 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 2.6 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.7 | GO:0070649 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 1.8 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 1.9 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 1.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.0 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.5 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 2.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 6.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 1.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.3 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 2.6 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.4 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 1.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 1.0 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.6 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 2.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0051836 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.5 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.8 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 1.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 1.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 4.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 3.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 1.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 5.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.9 | GO:0060077 | inhibitory synapse(GO:0060077) clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.1 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.6 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 2.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 4.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 1.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.4 | 3.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.3 | 3.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 4.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 2.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 2.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 0.9 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 0.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.6 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 4.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 4.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.5 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 2.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.3 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 1.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 4.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 1.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 1.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 1.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 2.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 2.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 3.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 2.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 4.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 4.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 4.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 3.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.7 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 0.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 2.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 5.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.3 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |